Figure 6.
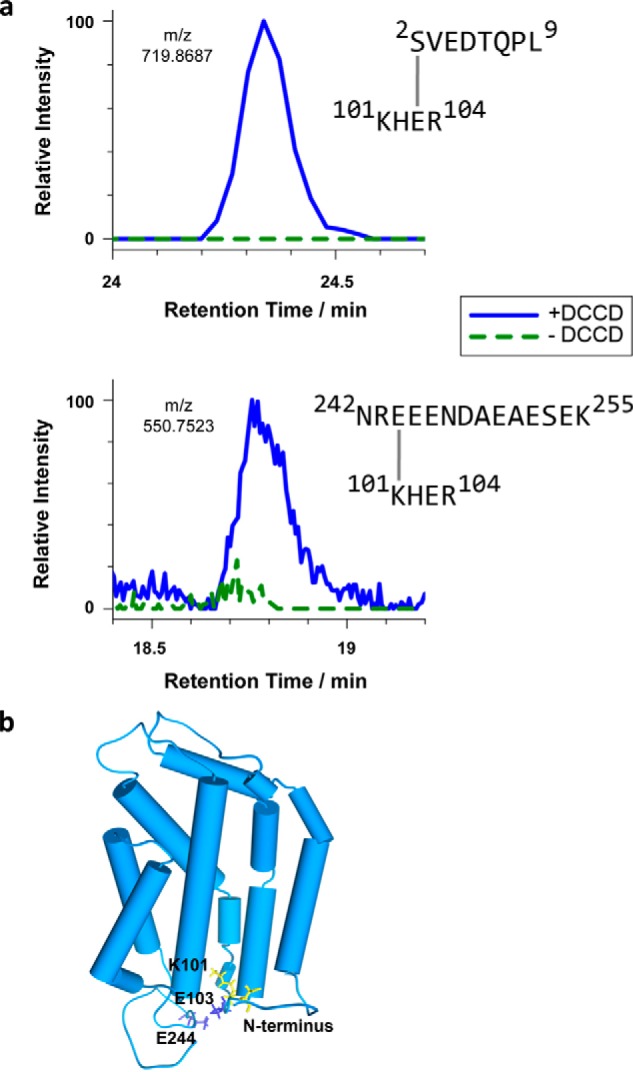
Intramolecular cross-linking by DCCD reveals conformational details of the RR-recognition site of TatC. a, extracted ion chromatograms of cross-linked TatC peptides analyzed by LC-MS. E. coli TatC treated with DCCD (blue curves) or mock-treated (dashed green curves) was isolated by affinity chromatography and SDS-PAGE prior to LC-MS. Identified cross-linked peptides represent intra-molecular TatC contacts between Ser2 and Glu103 (top), quantified on the 2+ charged precursor observed at m/z 719.8681 (molecular mass of 1437.7212 Da), and between Glu244 and Lys101 (bottom), quantified on the 4+ charged precursor at m/z 550.7523 (molecular mass of 2198.9788 Da). b, model of the E. coli TatC structure adapted from the crystal structure of A. aeolicus TatC (PDB codes 4B4A and 4HTT). Residues involved in the DCCD-mediated TatC cross-links specified in panel a are marked in blue (carboxylates) and yellow (amino groups). These intramolecular cross-links suggest an orientation of the N and C termini as modeled here, in which both cytosolic tails fold back toward the TM2/TM3 loop.
