Figure 1.
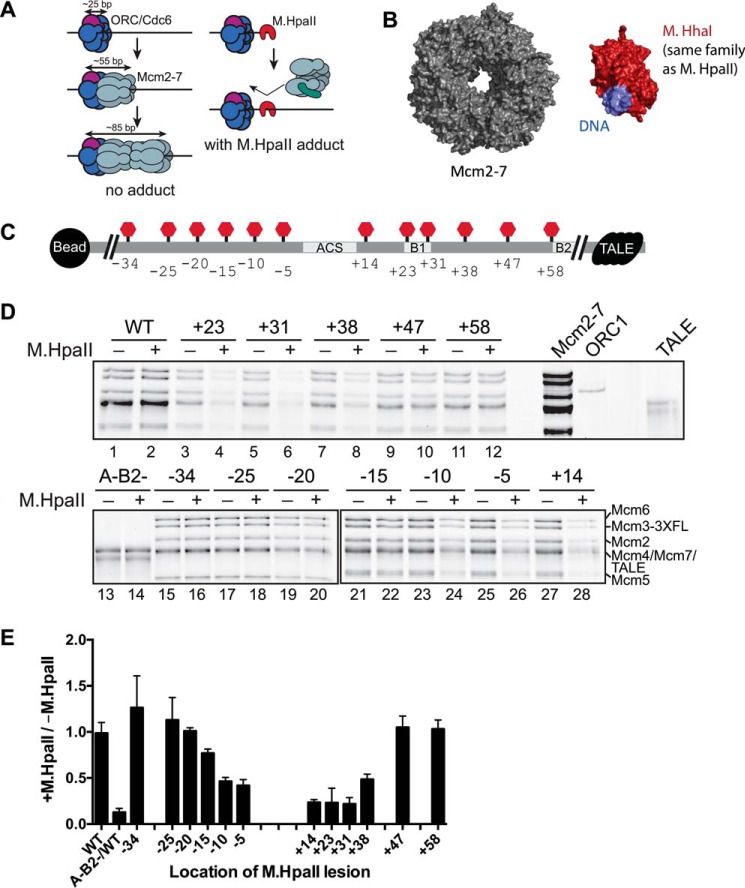
M.HpaII modification at a subset of locations within ARS1 affects Mcm2-7 helicase loading. A, rationale for using M.HpaII to inhibit helicase loading. Current models suggest ORC/Cdc6 binds to ∼25 bp of DNA, with the addition of each Mcm2-7 hexamer occupying an additional 30 bp of DNA. Binding of M.HpaII to a location in the origin DNA sequence where Mcm2-7 interacts should prevent stable helicase loading. B, structures of the Mcm2-7 ring (PDB code 3JA8) and a smaller but related S-adenosylmethionine-dependent methyltransferase (HhaI methyltransferase, PDB code 2Z6A) illustrate that the M.HpaII methyltransferase adduct is not expected to be accommodated within the Mcm2-7 central channel once the Mcm2-7 ring has closed around dsDNA. C, diagram showing the relative locations of the M.HpaII consensus sequence in the 12 different templates tested. Sites of M.HpaII cross-linking are shown as red “stop” signs (each template included only one modification). Changes at the origin sequence level are shown in Fig. S2A. Important origin sequences are indicated. All templates tested contained a TALE protein (black) bound to its consensus motif at the non-bead-attached end of the template to prevent helicases from sliding off the DNA. D, different M.HpaII-containing templates were assessed for their ability to load salt-stable Mcm2-7 complexes. Templates were either treated with M.HpaII (+) or left untreated (−). M.HpaII binding to 5FdC-modified DNA is quantitative (Fig. S2B). The location of the 5FdC-modified M.HpaII recognition motif is indicated above each pair of lanes. To eliminate any differences in DNA bead preparation or from introduction of the M.HpaII consensus sequence, relative changes between identical DNA templates either treated or untreated with M.HpaII were compared and quantified in E. E, comparative levels of Mcm2-7 loading across origins containing a 5FdC-modified M.HpaII recognition motif at the indicated location. Amount of helicase loading is reported as a fraction of loading when treated with M.HpaII relative to the level of loading on the same template without M.HpaII treatment. Error bars are the standard deviation from the mean calculated from three independent experiments.