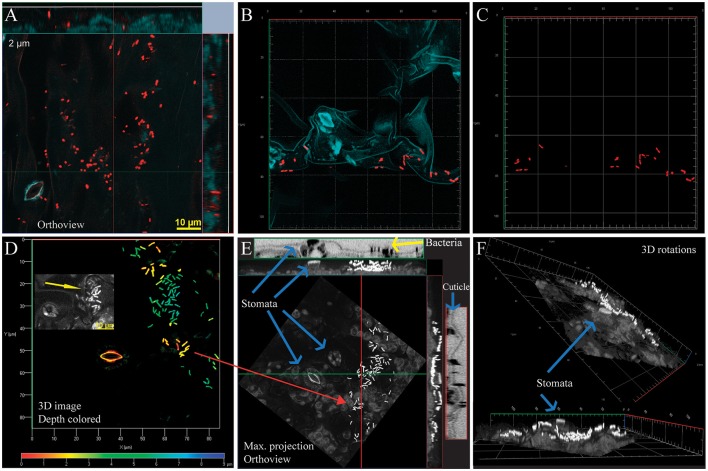
Figure 1.
Bacteria on the surface of greenhouse grown Arabidopsis associate strongly with leaf microstructures. All bacterial cells were labeled with the general probe EUB338 I, II, and III (here and elsewhere probe shown in red unless otherwise specified). All images were captured using a 100 X lens on a confocal microscope Zeiss LSM780. (A) Orthogonal projection, 13 confocal images. Bacteria on leaf surface distribute along the irregular profile of the leaf surface, as shown by the leaf autofluorescence (blue, here and elsewhere). (B) 3-D composite image using Z-stack capture, 6 images (5 μm depth), and analyzed using linear unmixing. (C) Same image as (B), background removed to facilitate visualization of bacteria. (D–F) Different views of the same image generated using Z-stack capture, 19 images (9 μm depth). (D) 3-D rendering image. The image is color-coded to indicate distance from the surface (red to dark blue). Inset shows high contrast detail to illustrate the cuticle irregularities in that area (yellow arrow). (E) Maximum projection orthoview (X, Y, and Z) showing bacteria on cuticle. Plant autofluorescence is shown in gray, bacteria in white. In the outside, orthoviews generated in Fiji from fluorescence data showcasing surface irregularities (blue arrows). Bacteria are shown in black (yellow arrow) (F) 3D renderings of same image. Further information can be found in Video S1 (fluorescence from the EUB338, transmitted light captured during confocal scanning microscopy, and overlap of both signals). Yellow bar, 10 μm.