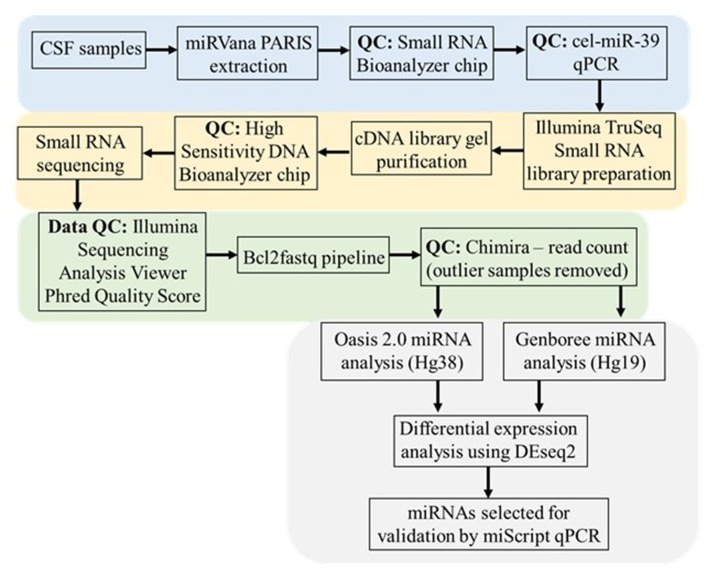
Figure 1.
CSF small RNA sequencing methodology and data analysis workflow. This workflow begins with RNA extraction from CSF samples using the miRVana PARIS RNA extraction kit. The proceeding quality control (QC) steps performed prior to small RNA sequencing include the running of a small RNA Bioanalyzer chip and qPCR for the cel-miR-39 spike-in. The extracted RNA was subjected to Illumina TruSeq library preparation and gel purification and subsequent QC steps prior to small RNA sequencing. Initial sequencing data was put through the Illumina Sequencing Analysis Viewer to establish the Phred Quality score (Q > 30). Following this the raw .bcl generated image files were de-multiplexed and converted to .fastq text files where initial QC testing was carried out in Chimira to establish the distribution of read counts amongst each sequenced sample and identify any outlier samples. Both Oasis 2.0 and Genboree platforms were used to identify the specific miRNA content in each sample prior to using DEseq2 in both platforms to determine differential expressed miRNAs in sALS patients compared to control subjects (p ≤ 0.05). Based on comparing the commonly expressed and uniquely significant miRNA between each analysis platform individual miRNAs were chosen for validation via miScript qPCR.