Abstract
Objective
This study investigated the association of yeast species with improved aerobic stability of total mixed ration (TMR) silages with prolonged ensiling, and clarified the characteristics of yeast species and their role during aerobic deterioration.
Methods
Whole crop corn (WCC) silages and TMR silages formulated with WCC were ensiled for 7, 14, 28, and 56 d and used for an aerobic stability test. Predominant yeast species were isolated from different periods and identified by sequencing analyses of the 26S rRNA gene D1/D2 domain. Characteristics (assimilation and tolerance) of the yeast species and their role during aerobic deterioration were investigated.
Results
In addition to species of Candida glabrata and Pichia kudriavzevii (P. kudriavzevii) previously isolated in WCC and TMR, Pichia manshurica (P. manshurica), Candida ethanolica (C. ethanolica), and Zygosaccharomyces bailii (Z. bailii) isolated at great frequency during deterioration, were capable of assimilating lactic or acetic acid and tolerant to acetic acid and might function more in deteriorating TMR silages at early fermentation (7 d and 14 d). With ensiling prolonged to 28 d, silages became more (p<0.01) stable when exposed to air, coinciding with the inhibition of yeast to below the detection limit. Species of P. manshurica that were predominant in deteriorating WCC silages were not detectable in TMR silages. In addition, the predominant yeast species of Z. bailii in deteriorating TMR silages at later fermentation (28 d and 56 d) were not observed in both WCC and WCC silages.
Conclusion
The inhibition of yeasts, particularly P. kudriavzevii, probably account for the improved aerobic stability of TMR silages at later fermentation. Fewer species seemed to be involved in aerobic deterioration of silages at later fermentation and Z. bailii was most likely to initiate the aerobic deterioration of TMR silages at later fermentation. The use of WCC in TMR might not influence the predominant yeast species during aerobic deterioration of TMR silages.
Keywords: Aerobic Stability, Deterioration, Silage, Total Mixed Ration, Yeast
INTRODUCTION
Aerobic deterioration is a critical problem for silage as it not only decreases the nutritional value but also affects animal or even human health [1,2]. To date numerous studies have been carried out on silages and indicated that aerobic deterioration is a microbial process carried out by aerobic microorganisms that cannot proliferate in the anaerobic environment of a sealed silo [3]. In well-preserved silage, the activities of microorganisms are greatly restricted by the rapid decline in pH and low final pH in addition to the anaerobic condition. Once the silo is opened, aeration will provide an opportunity for dormant aerobic microorganisms to multiply, and make the silages unstable. Yeasts are commonly reported to initiate aerobic deterioration of silage, as the oxidization of organic acids generally initiates aerobic spoilage [2,4].
Whole crop corn (WCC) silage is the main source of forage for lactating dairy cows, whereas, it deteriorated easily after exposure to air. There has been an increasing practice to ensile high-moisture by-products with dry feeds as fermented total mixed ration (TMR) silage in Japan. It has been indicated that materials with high moisture and water soluble carbohydrates (WSC) contents could be formulated as TMR silage and exhibit improved aerobic stability than ensiled alone [5,6]. As observed on corn silage [7], enhanced aerobic stability was concomitant with the decrease in yeast count. Similar results are also detected in our previous studies that decreased yeast were detected with prolonged ensiling, further improving the aerobic stability of TMR silages [5,8]. Furthermore, there are also evidences that yeast species might contribute more to aerobic deterioration than yeast count, as high stability could be obtained even when high populations (>104 colony forming unit [CFU]/g fresh matter [FM]) of yeast were detected [5]. Our previous research also indicated that yeasts are closely associated with the onset of the aerobic deterioration of TMR silage [6]. However, little research has focused on yeast dynamics during different periods and their characteristics associated with aerobic stability.
The objectives of this study were to investigate the predominant yeast species associated with prolonged ensiling and enhanced aerobic stability of TMR silages than single silage. In this study, both WCC and TMR inclusive of WCC were subjected to different ensiling periods (7, 14, 28, and 56 d) and used for aerobic stability test. We investigated the yeast dynamics and their characteristics associated with prolonged enisling and aerobic deterioration, and discussed the inclusive of WCC in yeast species and aerobic stability of TMR silages.
MATERIALS AND METHODS
Ensiling and aerobic stability test
Material of WCC (Zhongke 11, a high yield corn) was harvested in August 31 and at 198 g dry matter (DM)/kg of fresh forage, from Hebei Province located in China, chopped to approximately 2 to 3 cm in length, and then formulated with other by-products as TMR by a TMR mixer (Labrador, Storti, Verona, Italy). The formulation of TMR was mixed with WCC, soybean curd residue, soy sauce residue, corn starch residue, rice bran, corn meal, alfalfa, cotton meal, wheat bran, soybean meal, and a vitamin-mineral supplement at a ratio of 20:10:2.5:9:27:10:8:5:4:2:2.5, respectively, on a DM basis. After thoroughly mixing, WCC and TMR was immediately packed into high-density polyethylene barrels (60-L, 409 mm diameter and 619 mm height), compacted and sealed by a matched polyethylene cover with a gasket that was fixed by a steel circlip to avoid air infiltration. Triplicated barrels were randomly opened after 7, 14, 28, and 56 d of ensiling at ambient temperature (about 20°C to 25°C) and used for aerobic stability test.
After silo opening, the surface layers of silage were removed, and the rest were thoroughly scattered and then about 10 kg of sample was placed loosely back to a barrel without compaction. Non-fermented WCC and TMR were exposed to air immediately after preparation, following a similar procedure as that of silages. The barrels were covered by plastic lids to avoid drying but to allow air penetration. A thermocouple thermometer (Datataker, Victoria, Australia) was inserted into the core of sample to obtain temperature data. Changes in sample and room temperature were monitored every 10 min, and the records lasted for varied periods (from 5 to 25 d), depending on the extent of aerobic deterioration.
Aerobic stability was defined as the number of hours at which the silage remained stable before increasing to more than 2°C above room temperature. The sample was mixed every day after aerobic exposure and collected for analyses of yeast counts and predominant yeast species. Besides, yeast diversity of silages in three specific periods (the non-fermented materials, silages ensiled for 7, 14, 28, and 56 d at silo opening, and deteriorating silages (notable heat of 2°C above room temperature started to occur) for WCC and TMR (silages) were investigated.
Chemical component, pH and microbial count determination
Samples of WCC and TMR were taken immediately after thoroughly mixing, and silage samples were collected at silo opening and during aerobic exposure. The samples were dried in an oven at 60°C for 48 h, ground to pass through a 1-mm screen, and then used for chemical analysis. The DM content was analyzed according to method 934.01 of AOAC [9]. The WSC concentration was determined using the method of Owens et al [10]. The fermentation qualities were determined from water extracts of the silage [11]. The pH was measured using a glass electrode pH meter (S20K, Mettler Toledo, Zurich, Switzerland), and the organic acid content was determined by high performance liquid chromatography using the procedures described by Xu et al [11]. The numbers of microorganisms in fresh materials and silage samples were measured by plate count method [5]. Lactic acid bacteria were counted from de Man Rogosa Sharpe agar prepared using MRS broth (Difco, Detroit, MI, USA) with 1.6% agar, after incubation in an anaerobic incubator at 35°C for 3 d. Yeasts were enumerated on spread plates of Potato Dextrose Agar (Nissui, Tokyo, Japan) that had been acidified to pH 3.5 with sterilized tartaric acid. The plate cultures were incubated at 25°C for 3 to 5 d. The colonies were counted from the plates at appropriate dilutions and the number is expressed as CFU per gram of FM.
Isolation of yeast and extraction of genomic DNA
According to the colony morphology (texture, surface, elevation, margin, size, and color) and their frequency, about 10 representative yeast colonies were selected from Potato Dextrose Agar plates that were spread with appropriate dilutions of samples. The single and well-separated isolates were purified by repetitive streaking on yeast extract peptone dextrose agar (YEPD; containing 10 g/L yeast extract, 20 g/L bacteriological peptone, 20 g/L dextrose and 18 g/L agar). The purified isolates were stored in 20% glycerol at −80°C.
The genomic DNA of yeast was extracted according to the procedures that were described by Makimura et al [12], and the extracted genomic DNA of each strain was stored at −20°C until use.
26S rRNA gene D1/D2 domain sequencing
The D1/D2 domain of the 26S rRNA gene was amplified using the primer pair NL1 (5′-GCATATCAATAAGCGGAGGAAAAG -3′) and NL4 (5′-GGTCCGTGTTTCAAGACGG-3′) [13]. The DNA amplifications were carried out in a final volume of 50 μL containing 4 U of Taq DNA polymerase (Solarbio, Beijing, China), 1× polymerase chain reaction (PCR) reaction buffer (containing Mg2+), 0.2 mM each dNTP, 0.1 μM each primer and approximately 100 ng of template DNA. The PCR reaction was performed on a PCR thermal cycler (TP600, TaKaRa, Toyama, Japan) that was programmed as follows: 95°C for 5 min, followed by 36 cycles of denaturation at 94°C for 1 min, primer annealing at 52°C for 1 min, elongation at 72°C for 1 min 20 s, and finally elongation at 72°C for 8 min. The PCR products were separated by gel electrophoresis on a 1.0% agarose gel, detected by Gold View (Solarbio, Beijing, China) according to the manufacturer’s instructions and photographed under UV light with a charge-coupled device camera. The PCR products from the D1/D2 domain were directly sent to Sunny Biotechnology Co., Ltd. (Shanghai, China) for sequencing analysis.
Molecular phylogenetic analysis
Basic local alignment search tool (BLAST) searches of the sequences obtained were performed at the National Center for Biotechnology Information (NCBI) GenBank data library to determine the closest relatives of partial 26S rRNA gene sequences. The 26S rRNA gene sequences of isolates and sequences from the type strains held in GenBank were aligned with program CLUSTAL W [14] and adjusted manually. Phylogenetic trees were constructed from the evolutionary distance data that were calculated from Kimura’s two-parameter model [15] using the neighbor-joining method [16]. Bootstrap analyses [17] were performed on 1,000 random resamplings. Evolutionary analyses were conducted in MEGA 6 [18].
Trichosporon loubieri (T. loubieri) was used as an out-group organism. The nucleotide sequences for the 26S rRNA gene as described in this paper were deposited in the NCBI GenBank data library under accession numbers KM055439 to KM055471.
Microbial characteristic analysis
The pure cultures were cultivated at 30°C for 24 h before used for characteristic analysis. According to the silage pH, lactic or acetic acid assimilation by yeast was estimated by observing the pH change of modified YEPD medium (pH 4.0, 20 g/L of dextrose was replaced by 5 g/L lactic acid or 5 g/L acetic acid, respectively), after culturing at 30°C for 14 d. The significant increase in medium pH indicated a strain with strong lactic or acetic acid assimilation ability. Growth at 40°C was performed in YEPD broth after incubation in a thermostat water bath for 2 d. Growth at pH 3.5 was determined in YEPD broth after culturing at 30°C for 7 d. According to the lactic and acetic acid concentrations during ensiling, tolerance tests were set as 35 g/L of lactic acid or 8 g/L of acetic acid and the pH was adjusted to 4.0.
Data analysis
Microbial counts were log10-transformed to obtain normally distributed data and presented on a wet basis. The counts that below the detection limit (500 CFU/g FM) were assigned a value corresponding to half of the detection limit for subsequent statistical analysis. Data of fermentation characteristic and microbial count were analyzed with repeated measures, using the general linear model procedure of IBM SPSS Statistics for Windows (Version 20.0; IBM Corp., Armonk, NY, USA), through the following model: Yijk = μ+Si+Ej+(SE)ij+eijk, where Yijk is the observed value, μ is overall mean, Si is the effect associated with silage type, Ej is the effect associated with the ensiling periods, (SE)ij is the interaction effect between silage type and ensiling periods and eijk is residual. Duncan’s multiple range method was used to compare the means of different ensiling periods. The statistical significance was set as p<0.05.
RESULTS
Characteristics and aerobic stability of WCC and TMR silages
The WCC had high levels of WSC (28.5 g/kg FM) and moisture content (802 g/kg FM). After being incorporated into a TMR, the WSC and DM content was adjusted to 35.1 g/kg FM and 444 g/kg FM. After ensiling, the WCC and TMR silages were well-preserved, as indicated by a low pH of about 3.6 and 4.0, respectively. Non-fermented WCC and TMR deteriorated within 6 h of exposure. Whereas, silages exhibited greatly (p<0.01) enhanced aerobic stability with prolonged ensiling (Table 1, Figure 1). Besides, higher (p<0.01) aerobic stability was detected in TMR silage than that of WCC silage. After 28 d of ensiling, yeast was inhibited to below the detection limit (500 CFU/g FM).
Table 1.
Fermentation characteristic and microbial count of whole crop corn (WCC) and total mixed ration (TMR) silages during ensiling periods
| Items | pH | LA (g/kg FM) | AA (g/kg FM) | LAB (log10 CFU/g FM) | Yeast (log10 CFU/g FM) | Aerobic stability (h) |
|---|---|---|---|---|---|---|
| Silage type means | ||||||
| WCC | 4.24a | 18.07b | 5.28b | 7.40b | 3.88 | 67.1b |
| TMR | 4.08b | 28.37a | 5.96a | 8.02a | 3.83 | 192.7a |
| Ensiling period means (d) | ||||||
| 0 | 5.60a | 3.26d | 0.92d | 8.04b | 7.08a | 6.00e |
| 7 | 3.79c | 26.83c | 5.87c | 8.65a | 3.78b | 66.3d |
| 14 | 3.82b | 30.15a | 7.22a | 8.03b | 3.63b | 88.5c |
| 28 | 3.82b | 28.28b | 6.99b | 7.90b | 2.40c | 196.4b |
| 56 | 3.81bc | 27.57bc | 7.10ab | 5.94c | 2.40c | 292.2a |
| p-value | ||||||
| Silage type (S) | 0.037 | <0.001 | 0.002 | 0.004 | 0.598 | 0.002 |
| Ensiling periods (E) | 0.003 | <0.001 | <0.001 | 0.001 | 0.001 | 0.002 |
| Interaction (S×E) | 0.004 | 0.007 | 0.002 | <0.001 | 0.090 | 0.007 |
| SEM | 0.014 | 0.126 | 0.030 | 0.021 | 0.012 | 3.808 |
LA, lactic acid; AA, acetic acid; LAB, lactic acid bacteria; FM, fresh matter; CFU, colony forming unit; SEM, standard error of means; 2.40, i.e., log10 250 CFU/g, microbial count below the detection limit (500 CFU/g) is assigned a value corresponding to half of the detection limit, numerically for analysis of variance.
Means with different superscripts within columns significantly differed (p<0.05).
Figure 1.
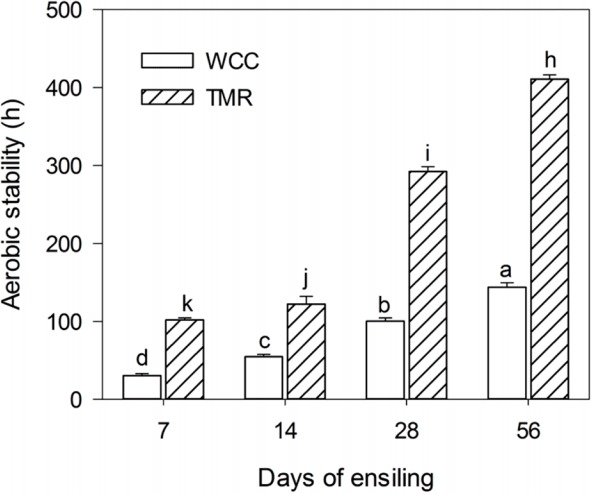
Aerobic stability of whole crop corn (WCC) and total mixed ration (TMR) silages ensiled for prolonged periods. Reported values represent the averages of triplicated analyses. Error bars represent the standard deviation. Bars with different superscript letters (a–d, h–k) are significantly different; p<0.05, when compared among the groups (Analysis of variance, followed by Duncan’s test).
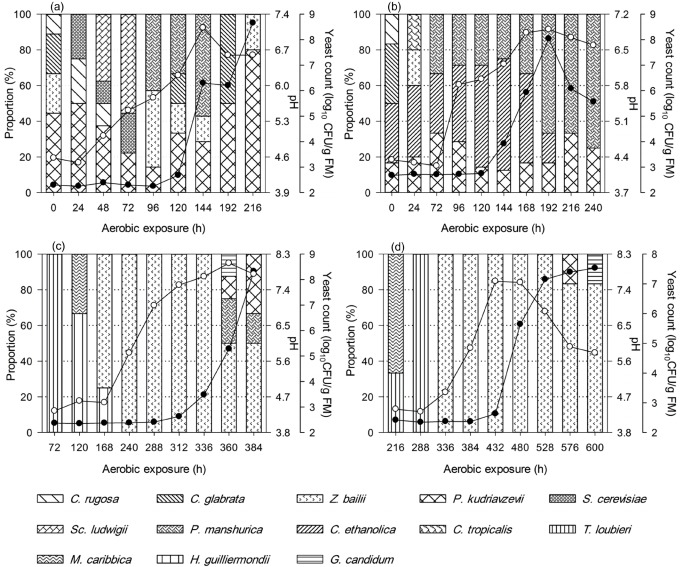
pH, yeast count and temperature dynamics of TMR silages during aerobic exposure
After exposure to air, pH and yeast count of non-fermented TMR decreased within 6 h of exposure (Figure 2). Whereas, TMR silages experienced a long and stable period in pH and temperature dynamics before heating, irrespective of ensiling periods (Figures 3, 4). Yeast started to propagate prior to the significant increase in pH and temperature. In addition, after exposed to air, the first temperature peak tended to be lower with the prolonged ensiling of TMR silages (Figure 4).
Figure 2.
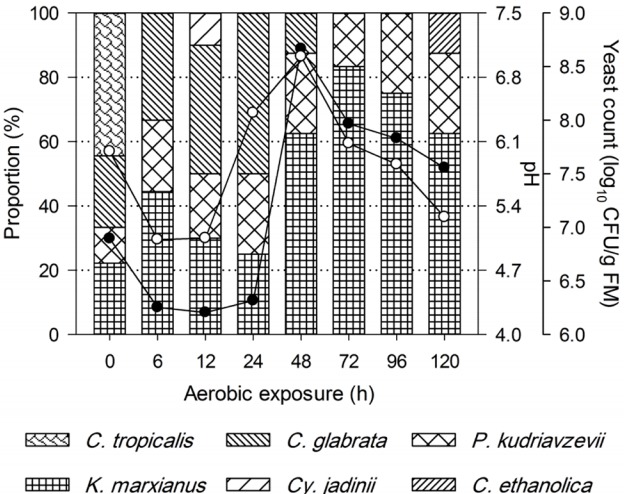
Changes in pH (●), yeast count (○) and yeast diversity during aerobic exposure of non-fermented total mixed ration (TMR).
Figure 3.
Changes in pH (●), yeast count (○) and yeast diversity during aerobic exposure of total mixed ration (TMR) silages that were ensiled for 7 d (a), 14 d (b), 28 d (c), and 56 d (d). Yeast counts below the detection limit (500 CFU/g) at the initial aerobic exposure period are not listed, which can be expressed as half of the detection limit (log10 250 CFU/g, i.e. 2.40). CFU, colony forming unit.
Figure 4.
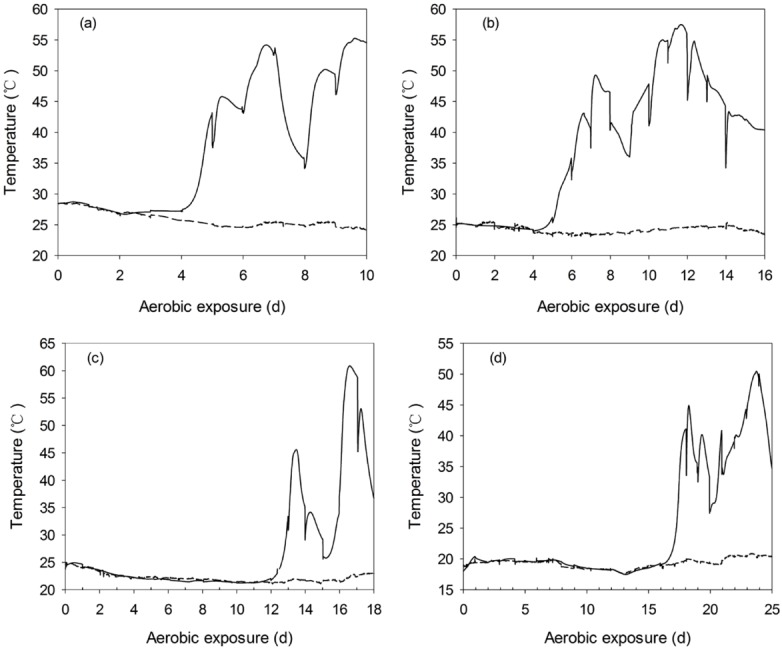
Changes of core temperature (—) and ambient temperature (---) during aerobic exposure of total mixed ration (TMR) silages that were ensiled for 7 d (a), 14 d (b), 28 d (c), and 56 d (d).
Yeast diversity associated with ensiling and aerobic deterioration of WCC and TMR silages
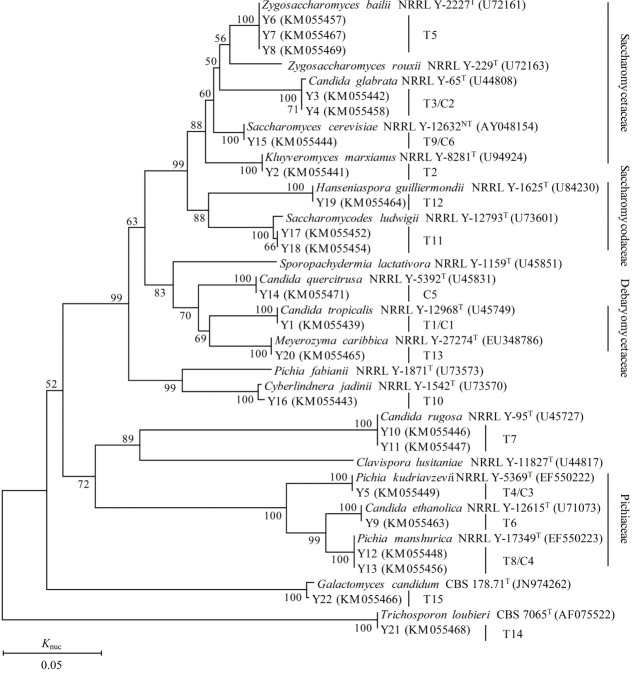
A total of 398 strains of yeast were isolated from TMR, TMR silages and aerobic exposed TMR silages, and 75 strains of yeast were isolated from WCC, WCC silages and deteriorating WCC silages. According to the 26S rRNA gene sequences, the isolates were identified as 16 yeast species and each strain was clearly assigned to related species. All of the strains were supported by 100% bootstrap values based on a bootstrap analysis of the phylogenetic tree and showing more than 97% similarity in 26S rRNA gene sequences with the corresponding type strains (Table 2, Figure 5). A total of 15 species (group T1–T15) were isolated from TMR (silages) and six species (group C1–C6) from WCC (silages). As shown in Figure 5, groups T2, T3/C2, T5, and T9/C6 were placed in clusters of Kluyveromyces marxianus (K. marxianus), Candida glabrata (C. glabrata), Zygosaccharomyces bailii and Saccharomyces cerevisiae (S. cerevisiae), respectively, belonging to Saccharomycetaceae. Groups of T11 and T12 were placed in clusters of Saccharomycodes ludwigii (Sc. ludwigii) and Hanseniaspora guilliermondii (H. guilliermondii), respectively, belonging to Saccharomycodaceae. T1/C1 and T13 were placed in clusters of Candida tropicalis (C. tropicalis) and Meyerozyma caribbica (M. caribbica), belonging to Debaryomycetaceae. T4/C3, T6, and T8/C4 were placed in clusters of Pichia kudriavzevii, Candida ethanolica (C. ethanolica), and P. manshurica, belonging to Pichiaceae. T7, T10, and T15 were assigned to clusters of Candida rugosa (C. rugose), Cyberlindnera jadinii (Cy. jadinii), and Galactomyces candidum (G. candidum), respectively. Group C5 was assigned to cluster of Candida quercitrusa (C. quercitrusa).
Table 2.
The proportion and characteristic of yeast species isolated from total mixed ration (TMR) and whole crop corn (WCC) and their silages subjected to different ensiling periods (7, 14, 28, and 56 d) during ensiling and after aerobic exposure
| Group | Similarity (%) | Reference strain (accession no.) | Proportion of yeast species isolated1) | Assimilation2) | Tolerance3) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||||
| TMR | E-T | D-T | WCC | E-C | D-C | LA | AA | pH | LA | AA | Temp | |||
| T1/C1 | 100 | Candida tropicalis (U45749) | 44.5 | nd | nd | 37.5 | 12.5 | nd | + | + | + | + | − | + |
| T2 | 100 | Kluyveromyces marxianus (U94924) | 22.2 | nd | nd | nd | nd | nd | + | w/− | + | + | − | + |
| T3/C2 | 99.7–99.8 | Candida glabrata (U44808) | 22.2 | 26.7 | 1.8 | 12.5 | 6.2 | 9.4 | + | + | + | + | + | + |
| T4/C3 | 100 | Pichia kudriavzevii (EF550222) | 11.1 | 33.4 | 8.9 | 12.5 | 43.8 | 3.3 | + | + | + | + | +/w | + |
| T5 | 99.3–100 | Zygosaccharomyces bailii (U72161) | nd | 13.3 | 57.1 | nd | nd | nd | − | +/w | + | + | +/w | − |
| T6 | 99.5–100 | Candida ethanolica (U71073) | nd | 13.3 | 16.1 | nd | nd | nd | + | + | + | + | + | − |
| T7 | 99.2–100 | Candida rugosa (U45727) | nd | 13.3 | nd | nd | nd | nd | + | + | + | + | + | + |
| T8/C4 | 99.8–100 | Pichia manshurica (EF550223) | nd | nd | 16.1 | nd | nd | 64.9 | + | + | + | + | + | W |
| C5 | 99.8 | Candida quercitrusa (U45831) | nd | nd | nd | 37.5 | nd | nd | + | + | + | + | − | − |
| T9/C6 | 100 | Saccharomyces cerevisiae (AY048154) | nd | nd | nd | nd | 37.5 | 22.4 | +/− | w/− | + | + | +/w | + |
| T10 | 97.9 | Cyberlindnera jadinii (U73570) | nd | nd | nd | nd | nd | nd | +/w | +/w | + | + | − | + |
| T11 | 99.0–99.3 | Saccharomycodes ludwigii (U73601) | nd | nd | nd | nd | nd | nd | w/− | w/− | + | + | w | − |
| T12 | 100 | Hanseniaspora guilliermondii (U84230) | nd | nd | nd | nd | nd | nd | − | − | + | + | − | − |
| T13 | 100 | Meyerozyma caribbica (EU348786) | nd | nd | nd | nd | nd | nd | w | w | + | + | − | + |
| T14 | 99.8–100 | Trichosporon loubieri (AF075522) | nd | nd | nd | nd | nd | nd | + | − | + | + | − | + |
| T15 | 98.3–98.5 | Galactomyces candidum (JN974262) | nd | nd | nd | nd | nd | nd | + | + | + | + | +/w | − |
TMR, fresh TMR; E-T, TMR silages during ensiling periods (7, 14, 28, and 56 d); D-T, deteriorating TMR silages (sample above room temperature by 2°C); WCC, fresh WCC; E-C, WCC silages during ensiling periods; D-C, deteriorating WCC silages.
Assimilation of lactic or acetic acid; LA, dextrose of YEPD medium was replaced by 5 g/L of lactic acid, pH 4.0; AA, dextrose of YEPD medium was replaced by 5 g/L of acetic acid, pH 4.0.
Tolerance test; pH, growth at YEPD medium (pH 3.5); LA, growth at YEPD medium with 35 g/L of lactic acid (pH 4.0); AA, growth at YEPD medium with 8 g/L of acetic acid (pH 4.0); Temp, growth test at 40°C.
nd, not detected;
+, positive (pH>4.20); w, weak (4.05<pH<4.20); −, negative (pH<4.05);
+, normal growth; w, weak growth; −, no growth.
Figure 5.
Phylogenetic tree drawn from neighbor-joining analysis of the 26S rRNA gene D1/D2 domain sequences, depicting the relationships of the isolated yeast species with closely related taxa. The bootstrap values for a total of 1,000 replicates are shown at the nodes of the tree. Trichosporon loubieri was used as an out-group. The bar indicates 5% sequence divergence. The GenBank accession no. of each type strain D1/D2 region 26S rRNA gene is shown behind the type strain. Knuc, nucleotide substitution rates.
Table 2 shows the frequency and characteristics of the above 16 different yeast species. A total of 10 species (eight from TMR and six from WCC) were detected from the three specific periods (non-fermented material, silages ensiled for 7, 14, 28, and 56 d and deteriorating silages). The other six species were detected after exposure but not included in the above three periods. For the four species isolated from non-fermented WCC, only species of P. kudriavzevii and C. glabrata were detectable during ensiling and deteriorating periods. P. kudriavzevii and S. cerevisiae were detected at high proportion in WCC silages. Whereas, P. manshurica was detected at the highest frequency in deteriorating WCC silages. No C. quercitrusa but K. marxianus was isolated after formulated as TMR. Species of C. glabrata and P. kudriavzevii were also detectable in deteriorating TMR silages and at high frequency in TMR silages. However, Z. bailii was predominant in deteriorating TMR silages.
Yeast dynamics during aerobic exposure of TMR and TMR silages
Figures 2 and 3 show the yeast dynamics of TMR and TMR silages after aerobic exposure. For the non-fermented TMR, few changes were detected in yeast species after exposure to air (Figure 2). C. tropicalis was exclusively found in fresh TMR, despite that yeast counts decreased during the initial 6 h of exposure, whereas K. marxianus, C. glabrata, and P. kudriavzevii were detected during the initial 48 h of exposure.
After 7 d of ensiling, C. tropicalis and K. marxianus became undetectable compared with TMR (Figure 3a). In addition to the species of C. glabrata and P. kudriavzevii that were previously detected in TMR, Z. bailii and C. rugosa were detected at silo opening. After exposure, species of S. cerevisiae and Sc. ludwigii were detected and absent within 96 h of exposure (before heat). P. manshurica started to be detected before heating and kept at a high frequency in deteriorating period. Besides, species of P. kudriavzevii, C. glabrata, and Z. bailii were also detected during heating. With ensiling prolonged to 14 d, C. ethanolica was isolated at silo opening, in addition to species of C. glabrata, P. kudriavzevii, and C. rugosa (Figure 3b). After exposed to air, C. ethanolica and P. kudriavzevii existed throughout aerobic exposure period. Besides, P. manshurica began to be detected at 72 h (before heat) and existed throughout the late period.
As ensiling prolonged to 28 and 56 d, the number of yeast decreased to below the detection limit at silo opening. Fewer yeast species were detected after exposed to air and the succession of yeast species was similar for the TMR silages at later fermentation (28 d and 56 d) (Figure 3c, 3d). T. loubieri, M. caribbica, and H. guilliermondii were occasionally detected before deterioration, as the yeast counts kept at or below the detection limit, and then for a long period, Z. bailii was the only yeast species that was found in aerobic-exposed and deteriorating silages. As deterioration proceeded, the pH increased and species of P. kudriavzevii, P. manshurica, and G. candidum became detectable.
DISCUSSION
In this study, both WCC and TMR silages were preserved well and TMR silages exhibited greater aerobic stability than WCC silages for the same ensiling period. These results are consistent with our previous studies [5]. Besides, the number of yeast decreased to below the detection limit, further improving aerobic stability. It agrees with our previous researches on TMR silages with different moisture contents [8].
In this study, 16 yeast species were isolated from non-fermented WCC and TMR, silages at silo opening and aerobic-exposed silages, mostly belonging to the phylum Ascomycota, except that T. loubieri belongs to the phylum Basidiomycota. Non-fermenting basidiomycetous yeast species were undetectable in either WCC or TMR, which were reported to vanish during ensiling and were replaced by ascomycetous species that were tolerant to acetic acid and could mostly vigorously ferment glucose [19]. In this study, except for T. loubieri, G. candidum, and P. manshurica, all of the detected species could vigorously ferment glucose [20]. Of the 16 yeast species, seven species belonging to C. quercitrusa, C. tropicalis, K. marxianus, Cy. jadinii, H. guilliermondii, M. caribbica, and T. loubieri can’t tolerate 8 g/L of acetic acid. This might explain their absence or less proportion during ensiling periods and the inhibited metabolic activity of H. guilliermondii, M. caribbica, and T. loubieri at the initial aerobic exposure of TMR silages at later fermentation.
It is well known that lactate-assimilating yeast (Candida and Pichia spp.) are generally the main initiators of the aerobic deterioration of silages, because of their acid tolerance and their metabolism of organic acids increases the pH thereby allowing the growth of less acid-tolerant microorganisms [1,4]. In this study, all the species that were previously detected in WCC and TMR are capable of assimilating lactic acid (Table 2). However, C. tropicalis and K. marxianus seemed to play fewer roles in TMR silages since they were undetectable during ensiling and after exposure, which might be attributable to the less tolerance of acetic acid. Z. bailii probably played lesser role in aerobic deterioration of TMR silages at early fermentation (7 d and 14 d), as indicated by the low detection frequency and incapability to assimilate lactic acid (Table 2, Figure 3). In this study, S. cerevisiae was detectable in WCC silages at silo opening and in deteriorating WCC silages. Whereas, for TMR silages, it was only detected in the 7-d TMR silage after 24 h of exposure. S. cerevisiae is considered a nonspoilage yeast because it does not assimilate lactic acid [21]. Jonsson and Pahlow [22] reported that the fermentative but non-lactate-assimilating S. cerevisiae prevailed under anaerobic conditions, whereas if air penetrated into the silage during storage, lactate-assimilating yeast of the genera Candida and Pichia predominated, as they have competitive advantage over S. cerevisiae under aerobic sugar-limited conditions. As shown in Table 2, some of the species S. cerevisiae isolated in this study are capable of assimilating lactic acid. Therefore, it probably involved in the deterioration of WCC silages, as indicated by the high proportion in deteriorating WCC silages. Besides, C. glabrata is reported to be lactate-tolerant and with high ability to grow under anaerobic conditions [23]. It has been isolated from spoiled corn silages [24]. In this study, C. glabrata and P. kudriavzevii seemed to involve in the aerobic deterioration of WCC silages and the 7-d TMR silages. Moreover, P. kudriavzevii was also detected in the 14-d TMR silages and existed after aerobic exposure. With ensiling prolonged to 28 and 56 d, aerobic stability was greatly enhanced and coincided with the inhibition of yeast to below the detection limit, including P. kudriavzevii. Therefore, the inhibition of yeast, particularly for P. kudriavzevii probably account for the enhanced aerobic stability of TMR silages at later fermentation. However, P. kudriavzevii was reported not seem to participate in aerobic deterioration, as it was detectable regardless of actual deterioration in Lactobacillus buchneri-inoculated WCC silage [24]. There are also evidences that yeast of the genus Pichia are usually the initial cause of aerobic deterioration in different silage crops [4] and previous research on corn silage also found that P. kudriavzevii was detected after aerobic exposure [25]. In addition, the improved aerobic stability in Lactobacillus buchneri-inoculated silage seemed to be associated with the inhibitory effect on P. anomala [26]. Sc. ludwigii has weak ability in assimilating organic acids and was exclusively detected before the deterioration of the 7-d TMR silage, therefore, might play lesser role in initiating deterioration. P. manshurica might play an important role in deterioration as it was detected at a high proportion in deteriorating WCC silages and TMR silages at early fermentation. Similar result was also found in sugar-cane silages at the beginning of aerobic exposure [27]. Besides, P. manshurica is commonly found in natural fermentation, but its role in spoilage is uncertain because of the past difficulty in resolving it from Pichia membranifaciens, which is regarded as a food and beverage spoilage organism [20]. With the prolonged ensiling, the inherent and spoilage yeasts were inhibited to below the detection limit, coincided with enhanced aerobic stability. Therefore, it is important to guarantee the sanitation of materials and TMR-related machinery to ensure less microbial contamination. In addition, a sufficient ensiling period and anaerobic conditions also contribute to the enhanced aerobic stability of TMR silages.
For the TMR silages at early fermentation, the role of yeast was difficult to explicate, as many yeast species were isolated during deteriorating period. In addition to the species that were detected at silo opening, S. cerevisiae, Sc. ludwigii, C. tropicalis, and P. manshurica were also detected after aerobic exposure. However, the former three species failed to be detected before heating. The notable heat of TMR silages at early fermentation was concomitant with a drastic increase in pH. Therefore, the deterioration of TMR silages at early fermentation probably due to the proliferation of P. manshurica, C. ethanolica, and P. kudriavzevii, which are capable of assimilating lactic acid.
With ensiling prolonged to 28 and 56 d, no yeast (<500 CFU/g FM) was detectable at silo opening and fewer species were detected after exposure. For the TMR silages at later fermentation, a small quantity (at or below the detection limit) of yeast belonging to T. loubieri, H. guilliermondii, and M. caribbica was occasionally and exclusively detected before heating. T. loubieri and M. caribbica were capable of assimilating lactic acid, whereas they failed to dominate deterioration and no significant increase in pH was detected. It probably due to the inhibited metabolic activity of T. loubieri and M. caribbica at the initial exposure period, as indicated by the occasionally and exclusively detection before heating and their less tolerance of acetic acid. However, Z. bailii was the only yeast species that was found during deteriorating period and capable of assimilating acetic acid. Therefore, Z. bailii was most likely to initiate the aerobic deterioration of TMR silages at later fermentation. Similar result was also obtained in our previous study on TMR silages with different moisture contents [8]. Besides the involvement of fewer yeast species, the incapable of assimilating lactic acid for species Z. bailii might also contribute to the reduced first temperature peak during heating. Although Z. bailii has no ability to utilize lactic acid, it possesses resistance to weak acid preservatives (e.g., acetic, benzoic and sorbic acids), osmo-tolerance, and the ability to vigorously ferment glucose as well as high tolerance of ethanol [28]. Furthermore, Z. bailii is one of the most common yeasts in fermentative food spoilage, particularly in food with a low pH and high sugar content [20]. There are also evidences that Z. bailii could grow in a medium with acetic acid, ethanol or glycerol as the sole carbon and energy source transported acetate by a saturable transport system [29]. This may account for the high resistance of Z. bailii to acidic media containing ethanol. The further utilization of acetic acid by Z. bailii or loss by volatilization will reduce the antifungal effect created by high acetic acid and facilitate the growth of other microorganisms.
In this study, species of P. kudriavzevii and C. glabrata were detected in both WCC and deteriorated WCC silages, however, not dominate in deteriorating TMR silages. Species P. manshurica that was predominant in deteriorating WCC silages was detectable exclusively in deteriorating TMR silages at early fermentation, however, not detected in TMR silages at later fermentation. In addition, the predominant yeast species of Z. bailii in deteriorating TMR silages at later fermentation was not detected in both WCC and WCC silages. Therefore, the inclusion of WCC in TMR in this study seemed not to influence the role of yeast species in deterioration of TMR silages.
According to previous studies, K. marxianus as well as Candida and Pichia species are the common spoilage organisms in dairy products for their ability to ferment lactose and utilize lactic acid [30]. In addition, the genera Candida, Pichia, Saccharomyces, and Zygosaccharomyces are potential spoilage yeasts in beer due to their resistance of alcohol, anaerobiosis, and low pH [31]. However, food and feed are, in general, very diverse microbial growth substrates. Different species have characteristics that make them more or less adapted to a particular system [32]. There is need to have a better understanding of the characteristics and role of yeast species in aerobic deterioration of TMR silages, so as to provide strategies to enhance aerobic stability. Culture dependent methods were used in this study to isolate the predominant yeast species and make it possible to investigate the characteristics of culturable yeast. This study enriches our understanding on culturable yeast associated with TMR silages and will aid in explore more suitable culture independent methods to strengthen the observations in the subsequent study.
To provide strategies to enhance aerobic stability, future work need to be carried on the mechanisms of aerobic stability of TMR silages as well as the characteristic analysis of deterioration-related species.
ACKNOWLEDGMENTS
This work was financially supported by the National Natural Science Foundation of China (31172240) and the National Scientific and Technological Support Projects (2017YFD0502102). The authors appreciate Pro. Bai Fengyan of the Systematic Mycology and Lichenology Laboratory, Institute of Microbiology, Chinese Academy of Sciences for technical assistance.
Footnotes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
REFERENCES
- 1.Driehuis F, Elferink SO. The impact of the quality of silage on animal health and food safety: a review. Vet Q. 2000;22:212–6. doi: 10.1080/01652176.2000.9695061. [DOI] [PubMed] [Google Scholar]
- 2.Woolford MK. The detrimental effects of air on silage. J Appl Bacteriol. 1990;68:101–16. doi: 10.1111/j.1365-2672.1990.tb02554.x. [DOI] [PubMed] [Google Scholar]
- 3.Courtin MG, Spoelstra SF. A simulation model of the microbiological and chemical changes accompanying the initial stage of aerobic deterioration of silage. Grass Forage Sci. 1990;45:153–65. [Google Scholar]
- 4.Pahlow G, Muck RE, Driehuis F, Elferink SJ, Spoelstra SF. Microbiology of ensiling. In: Buxton DR, Muck RE, Harrison JH, editors. Silage science and technology. Madison, WI, USA: American Society of Agronomy; 2003. pp. 31–93. [Google Scholar]
- 5.Hu X, Hao W, Wang H, et al. Fermentation characteristics and lactic acid bacteria succession of total mixed ration silages formulated with peach pomace. Asian-Australas J Anim Sci. 2015;28:502–10. doi: 10.5713/ajas.14.0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Ning T, Hao W, Zheng M, Xu C. Dynamics associated with prolonged ensiling and aerobic deterioration of total mixed ration silage containing whole crop corn. Asian-Australas J Anim Sci. 2016;29:62–72. doi: 10.5713/ajas.15.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabacco E, Piano S, Revello-Chion A, Borreani G. Effect of Lactobacillus buchneri LN4637 and Lactobacillus buchneri LN40177 on the aerobic stability, fermentation products, and microbial populations of corn silage under farm conditions. J Dairy Sci. 2011;94:5589–98. doi: 10.3168/jds.2011-4286. [DOI] [PubMed] [Google Scholar]
- 8.Hao W, Wang H, Ning T, Yang F, Xu C. Aerobic stability and effects of yeasts during deterioration of non-fermented and fermented total mixed ration with different moisture levels. Asian-Australas J Anim Sci. 2015;28:816–26. doi: 10.5713/ajas.14.0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.AOAC . Offcial Methods of Analysis. 15th ed. Arlington, VA, USA: Association of Official Analytical Chemists; 1990. [Google Scholar]
- 10.Owens V, Albrecht K, Muck R, Duke S. Protein degradation and fermentation characteristics of red clover and alfalfa silage harvested with varying levels of total nonstructural carbohydrates. Crop Sci. 1999;39:1873–80. [Google Scholar]
- 11.Xu C, Cai Y, Moriya N, Ogawa M. Nutritive value for ruminants of green tea grounds as a replacement of brewers’ grains in totally mixed ration silage. Anim Feed Sci Technol. 2007;138:228–38. [Google Scholar]
- 12.Makimura K, Murayama SY, Yamaguchi H. Detection of a wide range of medically important fungi by the polymerase chain reaction. J Med Microbiol. 1994;40:358–64. doi: 10.1099/00222615-40-5-358. [DOI] [PubMed] [Google Scholar]
- 13.Kurtzman CP, Robnett CJ. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek. 1998;73:331–71. doi: 10.1023/a:1001761008817. [DOI] [PubMed] [Google Scholar]
- 14.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 16.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 18.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Middelhoven WJ, van Baalen AH. Development of the yeast flora of whole-crop maize during ensiling and during subsequent aerobiosis. J Sci Food Agric. 1988;42:199–207. [Google Scholar]
- 20.Kurtzman C, Fell JW, Boekhout T. The yeasts: a taxonomic study. London, UK: Elsevier; 2011. [Google Scholar]
- 21.McDonald P, Henderson A, Heron S. The biochemistry of silage. 2nd ed. Welton, Lincoln, UK: Chalcombe Publications; 1991. [Google Scholar]
- 22.Jonsson A, Pahlow G. Systematic classification and biochemical characterization of yeasts growing in grass silage inoculated with Lactobacillus cultures. Anim Res Dev. 1984;20:7–22. [Google Scholar]
- 23.Merico A, Sulo P, Piškur J, Compagno C. Fermentative lifestyle in yeasts belonging to the Saccharomyces complex. FEBS J. 2007;274:976–89. doi: 10.1111/j.1742-4658.2007.05645.x. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Nishino N. Effects of inoculation of Lactobacillus rhamnosus and Lactobacillus buchneri on fermentation, aerobic stability and microbial communities in whole crop corn silage. Grassl Sci. 2011;57:184–91. [Google Scholar]
- 25.Dolci P, Tabacco E, Cocolin L, Borreani G. Microbial dynamics during aerobic exposure of corn silage stored under oxygen barrier or polyethylene films. Appl Environ Microbiol. 2011;77:7499–507. doi: 10.1128/AEM.05050-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Nishino N. Bacterial and fungal communities of wilted Italian ryegrass silage inoculated with and without Lactobacillus rhamnosus or Lactobacillus buchneri . Lett Appl Microbiol. 2011;52:314–21. doi: 10.1111/j.1472-765X.2010.03000.x. [DOI] [PubMed] [Google Scholar]
- 27.Carvalho B, Ávila C, Miguel M, et al. Aerobic stability of sugar-cane silage inoculated with tropical strains of lactic acid bacteria. Grass Forage Sci. 2015;70:308–23. [Google Scholar]
- 28.Kalathenos P, Sutherland J, Roberts T. Resistance of some wine spoilage yeasts to combinations of ethanol and acids present in wine. J Appl Bacteriol. 1995;78:245–50. [Google Scholar]
- 29.Sousa MJ, Rodrigues F, Coôrte-Real M, Leão C. Mechanisms underlying the transport and intracellular metabolism of acetic acid in the presence of glucose in the yeast Zygosaccharomyces bailii . Microbiol. 1998;144:665–70. doi: 10.1099/00221287-144-3-665. [DOI] [PubMed] [Google Scholar]
- 30.Pitt JI, Hocking AD, Diane A. Fungi and food spoilage. 3rd ed. New York, USA: Springer; 2009. [Google Scholar]
- 31.Campbell I. Beer. In: Doyle MP, Beuchat LR, editors. Food microbiology: fundamentals and frontiers. 3rd ed. Washington, DC, USA: ASM Press; 2007. pp. 851–62. [Google Scholar]
- 32.Ström K. Fungal inhibitory lactic acid bacteria. Uppsala, Sweden: Swedish University of Agricultural Sciences; 2005. [Google Scholar]