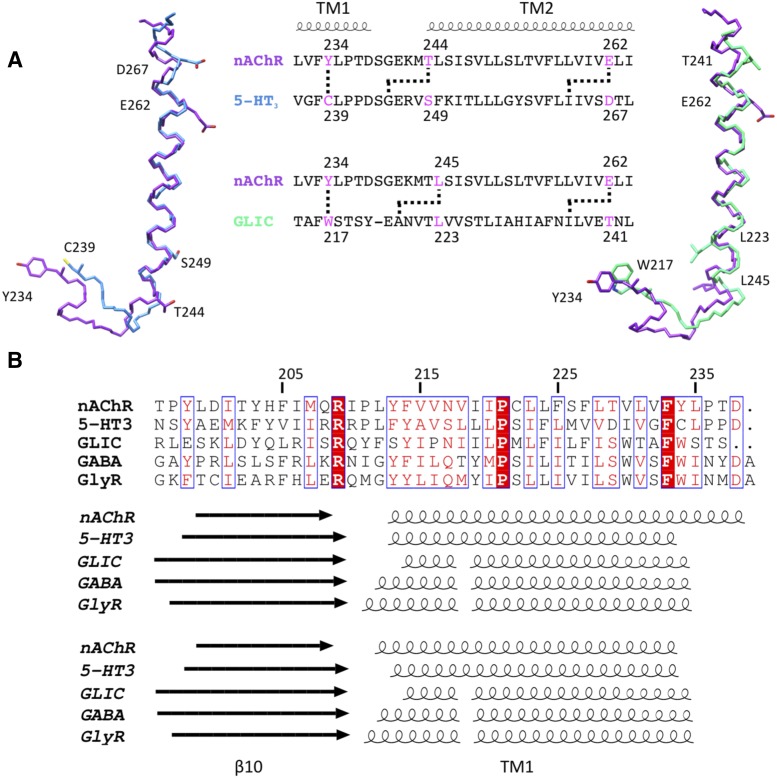
Fig. 8.
Refinement of the transmembrane domain of the Torpedo nAChR αγ subunit. (A) Sequence alignment and structural superposition of the TM1-TM2 loop region and TM2 helix of the Torpedo αγ subunit (PDB ID 2BG9; chain A) with that of the 5-HT3 receptor (PDB ID 4PIR; chain A) and GLIC (PDB ID 4HFI; chain A). As has been described previously (Supplemental Fig. 21 in Hibbs and Gouaux, 2011), amino acids from the TM1-TM2 loop of the Torpedo nAChR (e.g., Y234) superpose well with homologous amino acids from other pLGIC structures (C239 of 5-HT3R and W217 of GLIC; indicated by a straight line in the sequence alignment). In contrast, amino acids within the nAChR TM2 domain are out of register by ∼1 turn of the α-helix when compared with other pLGIC structures (e.g., compare E262 of the nAChR structure with D267 of 5-HT3R and T241 of GLIC; indicated by an angled line in the sequence alignment). (B) Alignment of amino acid sequence of the β10 strand and TM1 helix of Torpedo nAChR α subunit with that of related pLGICs (top panel). Also shown is the secondary structure before refinement (middle panel) and after refinement (bottom panel) of the Torpedo nAChR structure. Structural information is derived from the following PDB files: nAChR (2BG9; chain A), 5-HT3 receptor (4PIR; chain A), GABAA receptor (4COF; chain A), glycine receptor (3JAD; chain A), and GLIC (4HFI; chain A). Arrows denote β-strands, spirals denote α-helices, conserved residues are highlighted with white text on a red background, and residues with similar properties are highlighted with red text on a white background.