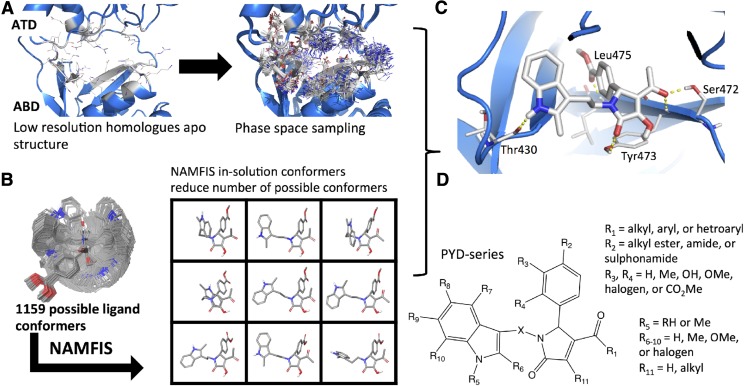
Fig. 2.
An overview of how a bioactive ligand-receptor complex was selected. (A) The receptor-based modeling highlights how homology models were generated from low-resolution crystal structure (3.59–3.96 Å) to capture possible conformations of the active site. (B) The ligand-based portion shows how in-solution conformers were determined using NMR-derived constraints and NAMFIS. (C) The ligand-receptor complex part illustrates that various different techniques including quantitative structure–activity relationship, molecular dynamics and docking were used to select a representative bioactive ligand-receptor complex. (D) The PYD series highlights the SAR developed by Zimmerman et al. (2014).