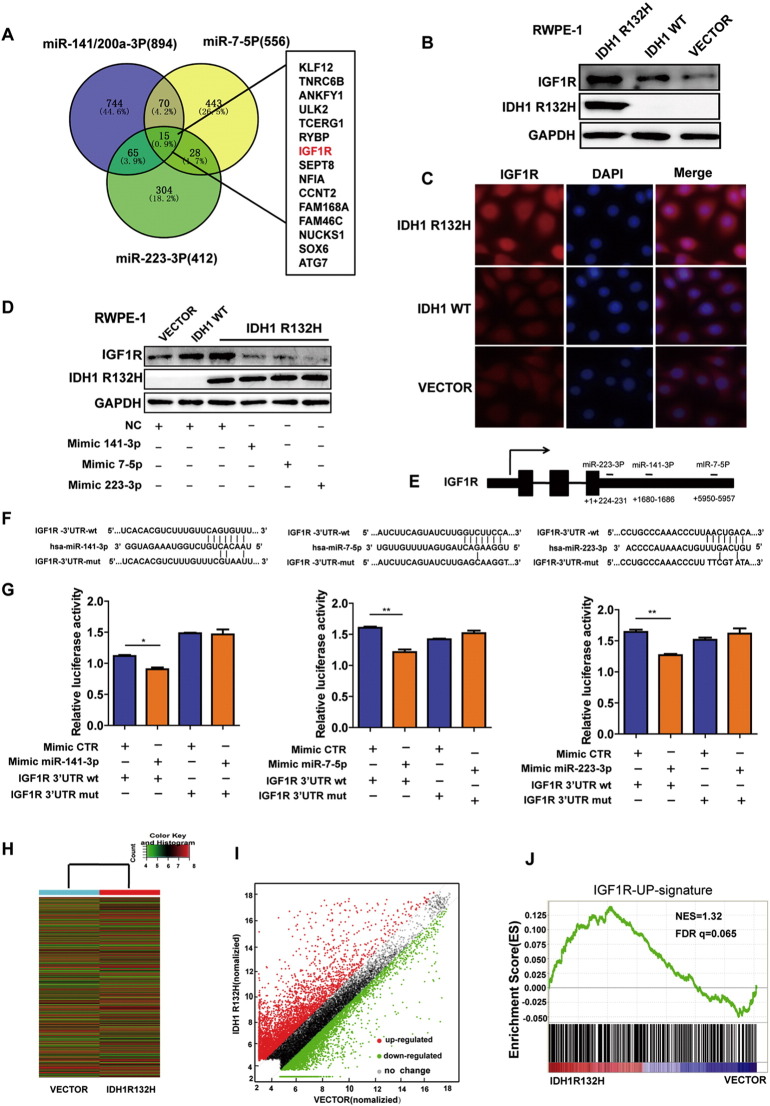
Figure 4.
IDH1R132H-downregulated miRNAs lead to increased IGF1R expression. (A) The predicted target gene analysis of miR-141-3p, miR-223-3p, and miR-7-5p was performed by TargetScan software. (B, C) Western blot and immunofluorescence were used to examine IGF1R protein levels in RWPE-1 cells expressing IDH1R132H, IDH1WT, and VECTOR. (D) The protein levels of IGF1R were assessed in IDH1R132H-mutant RWPE-1 cells after treatment with miR-141-3p, miR-223-3p, and miR-7-5p mimics or negative control (Western blot). (E) Diagram indicates the recognition sites for miR-141-3p, miR-223-3p, and miR-7-5p in IGF1R 3′-UTR region, respectively. (F) WT- and mutated recognition sites of miR-141-3p, miR-223-3p, and miR-7-5p in IGF1R 3′-UTR region. (G) LUC reporter assay was performed in HEK293T cells. Cells were transiently co-transfected with wt- or mutated IGF1R 3′-UTR region together with corresponding miRNA mimics, respectively. After incubation for 48 hours, luciferase activities were measured. (H, I) Heat map and scatter diagram were used to indicate the different gene expression of IDH1R132H and VECTOR in RWPE-1 cells. (G) IGF1R-upregulated (GSE5225) gene signatures were further analyzed by GSEA in mRNA microarray of RWPE-1 cells expressing IDH1R132H and VECTOR. FDR q = 0.065. All histograms represent mean ± SD of at least three independent replicates. P values: *P < .05, **P < .01.