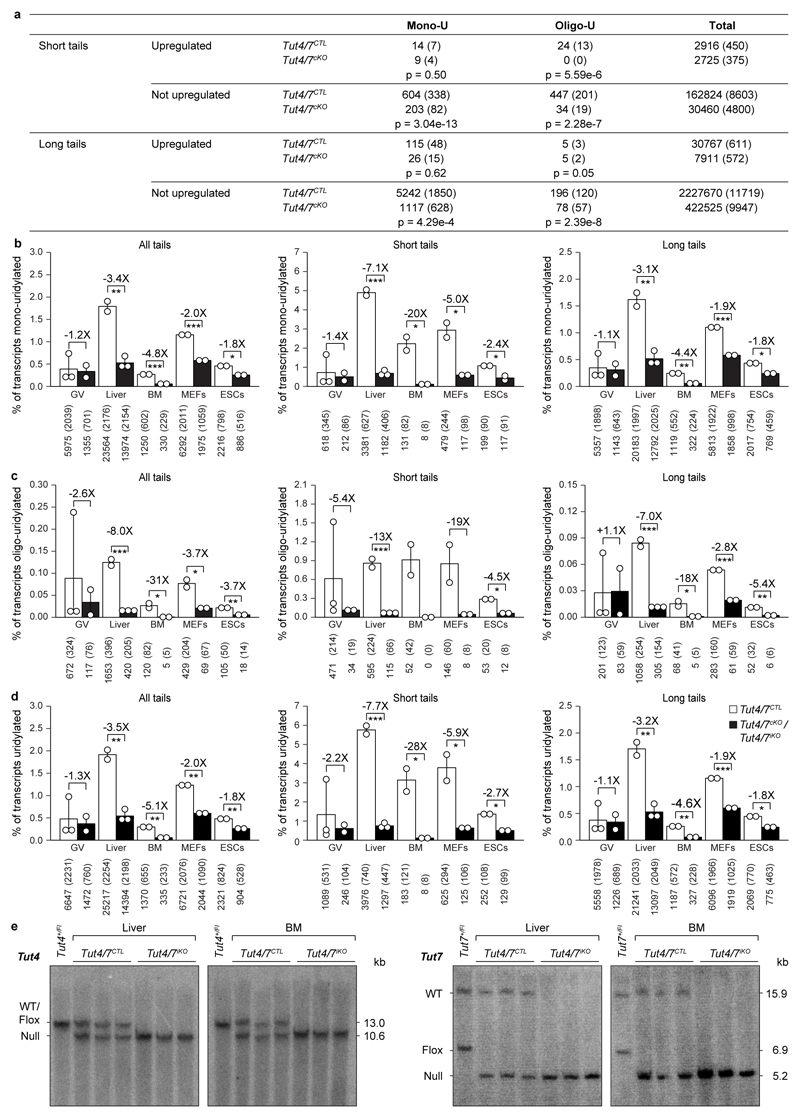
Extended Data Figure 7. Loss of TUT4/7 reduces both mono- and oligo-uridylation in different tissues and cell lines.
a, Transcript count for upregulated and not upregulated genes in Tut4/7CTL and Tut4/7cKO GV oocytes. The number of transcripts and in brackets the number of genes represented by those transcripts is presented. The p-value determined using the Pearson's chi-squared test is shown. b-d, Quantification of mRNA 3′ terminal mono- (b), oligo- (c) and total uridylation (d) of all transcripts (left panel) as well as those with short (≤ 30 nucleotides) (center panel) and long (> 30 nucleotides) (right panel) poly(A) tails of Tut4/7CTL and Tut4/7cKO germinal vesicle (GV) oocytes as well as Tut4/7CTL and Tut4/7iKO liver, bone marrow (BM), mouse embryonic fibroblasts (MEFs) and embryonic stem cells (ESCs). Data points represent replicates; the bars’ heights and vertical lines indicate the mean and range, respectively. The fold reduction in uridylation and the significance are indicated (t-test; *, p < 0.05; **, p < 0.01; ***, p < 0.001). e, Southern blots of EcoRV-digested genomic DNA from Tut4/7CTL and Tut4/7iKO liver and bone marrow (BM) confirming deletion of Tut4 and Tut7 after tamoxifen administration. The expected sizes for the wild type (WT), the conditional (flox) and the null alleles are indicated.