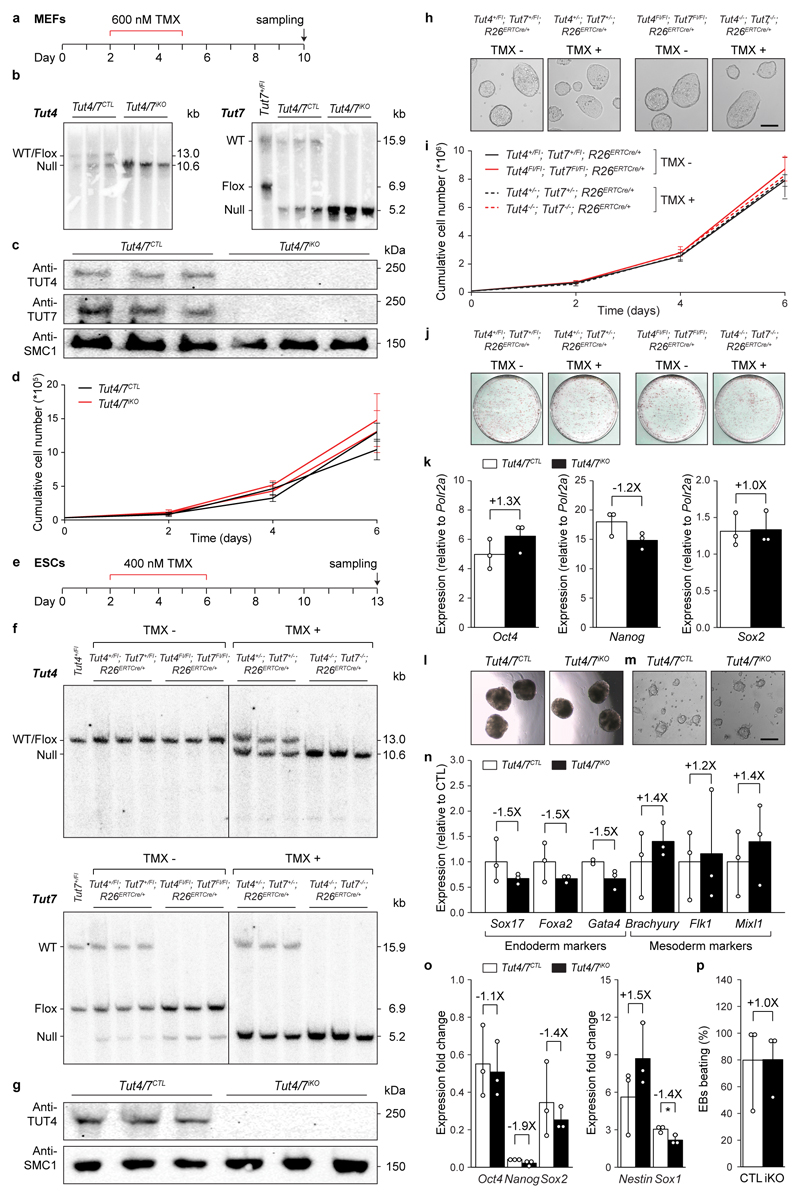
Extended Data Figure 8. Deletion of Tut4/7 in mouse embryonic fibroblasts and stem cells.
a, Strategy and timing for the acute deletion of Tut4/7 in mouse embryonic fibroblasts (MEFs). TMX denotes 4-OH-tamoxifen. b, Southern blots of EcoRV-digested genomic DNA from Tut4/7CTL and Tut4/7iKO MEF cell lines confirming deletion of Tut4 and Tut7. The expected sizes for the wild type (WT), the conditional (flox) and the null alleles are indicated. c, Western blots probed with the indicated antibodies from Tut4/7CTL and Tut4/7iKO MEF whole cell extracts. d, Growth curve of two independent Tut4/7CTL and Tut4/7iKO MEF cell lines. The mean and s.d. of three technical replicates is shown. e, Strategy and timing for the acute deletion of Tut4/7 in mouse embryonic stem cells (ESCs). f, Southern blots of EcoRV-digested genomic DNA from Tut4+/Fl; Tut7+/Fl; R26ERTCre/+ and Tut4Fl/Fl; Tut7Fl/Fl; R26ERTCre/+ ESC lines before (TMX-) and after (TMX+) deletion of Tut4/7. The expected sizes for the wild type (WT), the conditional (flox) and the null alleles are indicated. g, Western blots probed with the indicated antibodies from Tut4/7CTL and Tut4/7iKO ESC whole cell extracts. h, Representative images showing morphology of Tut4+/Fl; Tut7+/Fl; R26ERTCre/+ and Tut4Fl/Fl; Tut7Fl/Fl; R26ERTCre/+ ESC lines before (TMX-) and after (TMX+) deletion of Tut4/7. Scale bar indicates 200 μm. i, Growth curve of Tut4+/Fl; Tut7+/Fl; R26ERTCre/+ and Tut4Fl/Fl; Tut7Fl/Fl; R26ERTCre/+ESC lines before (TMX-) and after (TMX+) deletion of Tut4/7. The growth of each line was measured in triplicates. The mean and s.d. of three independently-derived cell lines is shown. j, Alkaline phosphatase staining of untreated (TMX-) and tamoxifen treated (TMX+) Tut4+/Fl; Tut7+/Fl; R26ERTCre/+ and Tut4Fl/Fl; Tut7Fl/Fl; R26ERTCre/+ ESC lines. k, Relative expression of pluripotency markers in Tut4/7CTL and Tut4/7iKO ESC lines as determined by qRT-PCR from three biological replicates. The bars’ heights and vertical lines indicate the mean and range, respectively. The fold change is indicated. l, Representative images showing morphology of Tut4/7CTL and Tut4/7iKO embryoid bodies. Magnification, 20X. m, Representative images showing morphology of neural progenitors derived from of Tut4/7CTL and Tut4/7iKO ESCs. Scale bar indicates 100 μm. n, Relative expression as determined by qRT-PCR of endoderm and mesoderm markers in embryoid bodies derived from Tut4/7CTL and Tut4/7iKO ESCs. The mean from three biological replicates is shown. The bars’ heights and vertical lines indicate the mean and range, respectively. The fold change is indicated. o, Expression as determined by qRT-PCR presented as fold change of pluripotency and neural progenitor markers upon differentiation of Tut4/7CTL and Tut4/7iKO ECSs into neural progenitors. The fold change is indicated. The bars’ heights and vertical lines indicate the mean and range, respectively (t-test; *, p < 0.05). p, Frequency of Tut4/7CTL and Tut4/7iKO embryoid bodies with spontaneous contractile activity. The mean from three biological replicates is shown, each determined from 48 embryoid bodies. The fold change is indicated. The bars’ heights and vertical lines indicate the mean and range, respectively.