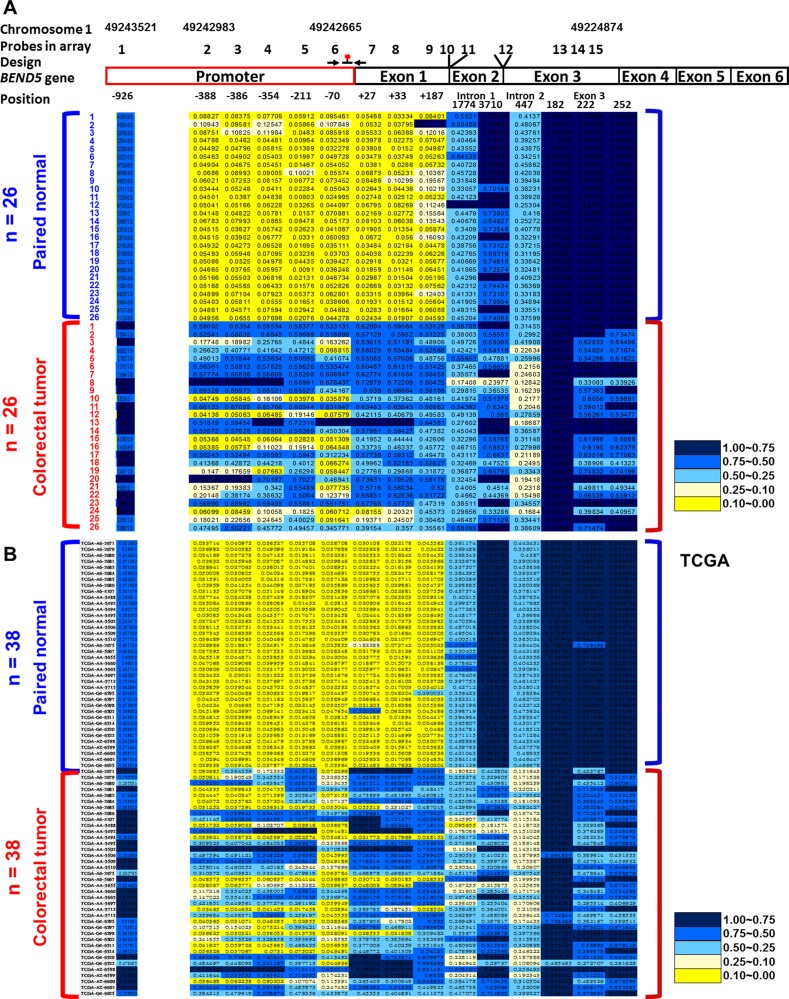
Figure 1. Differentially methylated CpG in BEND5 in CRC patients.
Methylation levels (average β values) at the differentially methylated loci were identified using an Illumina Methylation 450K array-based assay in (A) 26 CRC patients in Taiwan and (B) 38 CRC patients from the TCGA data set. The scale shows the relative methylation status from 0.00 to 1.00 (yellow: hypomethylation, blue: hypermethylation). Fifteen CpG sites on BEND5 were detected in 26 paired normal (upper) and CRC (lower) tissues. Six CpG sites in promoter regions −926, −388, −386, −354, −211, and −70 are designated 1, 2, 3, 4, 5, and 6, respectively. The CpG sites in exon 1 regions +27, +33, and +187 are designated 7, 8, and 9, respectively. The 1774 and 3710 sites in intron 1 regions are designated 10 and 11, respectively. One site is in the intron 2 region (447) and designated 12. Three CpG sites in exon 3 regions 182, 222, and 252 are designated 13, 14, and 15, respectively. The BEND5 gene is located on chromosome 1. Positions 1, 2, 6, and 15 are on 49243521, 49242983, 49242665, and 49224874 of chromosomes 1, respectively. Primers and the probe for the QMSP assay are marked and indicated in the junction between the promoter and exon 1 regions.