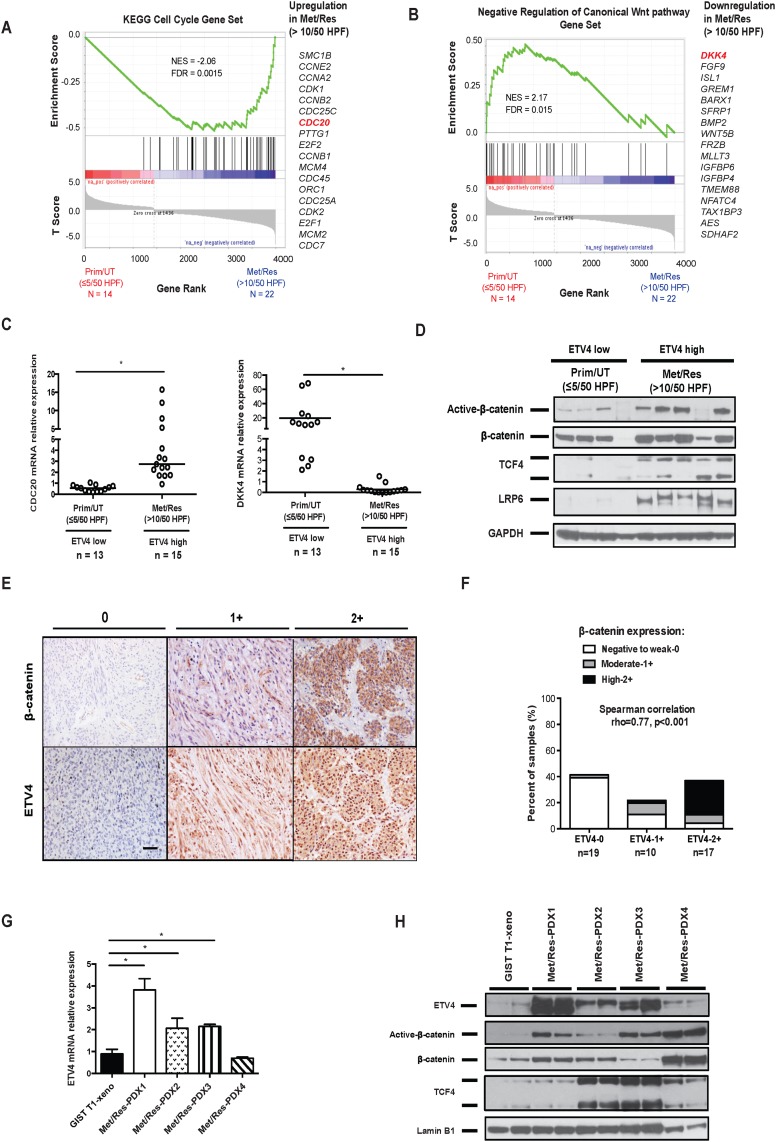
Figure 6. ETV4 upregulation is associated with canonical Wnt activation in human GISTs.
GSEA was performed on high throughput RNA sequencing data from 36 selected human GISTs. (A) Enrichment of cell cycle genes and (B) negative regulation of Wnt genes was found in Met/Res (>10/50 HPF) GISTs (N = 22) compared to Prim/UT (<5/50 HPF) GISTs (N = 14). False Discovery Rate (FDR) and Normalized Enrichment Score (NES) were indicated. (C) The relative mRNA expression of CDC20 and DKK4 was analyzed by real-time PCR in ETV4-low and high GIST tumors. Horizontal bars represent the median. Prim/UT (<5/50 HPF) – primary, untreated GIST (n = 13), Met/Res (>5/50 HPF) – metastatic, imatinib-resistant GIST (n = 15). Student’s t test; *P < 0.05. (D) Western blot of Wnt pathway components in ETV4-high (metastatic, imatinib-resistant GISTs (Met/Res) with a mitotic rate >10/50 HPF) or ETV4-low (primary, untreated GISTs (Prim/UT) with a mitotic rate ≤5/50 HPF). (E) Representative staining of β-catenin and ETV4 expression in 46 GIST specimens. Staining intensity was scored as negative to weak (0), moderate (1+), or high (2+). Scale bar, 20 μm. (F) Correlation of ETV4 and β-catenin expression staining in 46 GISTs. Spearman’s rho = 0.77 with P = 0.001 per 2-tailed t test. (G) Real-time PCR showing relative mRNA expression of ETV4 in four PDXs compared to GIST T1 xenografts. All bars, means ± SEM. Student’s t test; *P < 0.05. (H) Immunoblots of nuclear extracts from GIST T1 xenografts and four PDXs. PDX-1: KIT exon 11/13 mutation; PDX-2: WT KIT with SDHB/KRAS mutation; PDX-3: KIT exon 9/11 mutation; PDX-4: KIT exon 11 mutation with CDKN2A deletion. All four PDXs are from tumors of metastatic, imatinib-resistant GIST patients with a high mitotic rate.