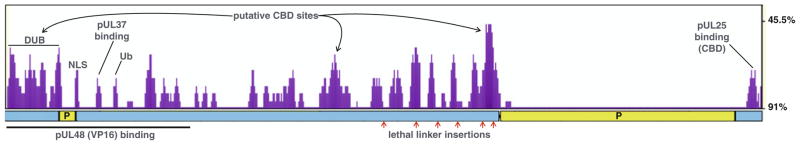
Fig. 8.4.
VP1/2 amino acid conservation plot. The plot of cumulative divergence indicates regions of the large tegument protein that are predicted functional elements under selective pressure. Evolutionary stable regions of the protein are marked by the higher peaks (purple). Overlaid on the plot are annotations of protein regions discussed in this review, including: the deubiquitinase (DUB), pUL37 binding site; ubiquitin switch site (Ub), and the C-terminal capsid binding domain (CBD). A nuclear localization signal (NLS) that is critical for infection is also indicated (Abaitua and O’Hare 2008; Abaitua et al. 2012). The locations of three additional proposed CBD’s are also indicated. Below the plot is a linear illustration of VP1/2 highlighting the two regions that are rich in proline (P) and are predicted to be disordered regions of the protein, and below this are annotations for the region of VP1/2 that interacts with VP16 by yeast-2-hybrid analysis, and six linker insertion sites that are lethal to PRV (Mohl 2010). The plot was produced with VP1/2 amino acid sequences from seven alpha-herpesviruses (BHV-1, EHV-1, EHV-4, HSV-1, HSV-2, PRV, and VZV) using a sliding window size of 25 on the eShadow web site: eshadow.dcode.org (Ovcharenko 2004)