Abstract
The last few decades have seen the development of many high performance separation methods that use biologically-related binding agents. The combination of HPLC with these binding agents results in a technique known as high performance affinity chromatography (HPAC). This review will discuss the general principles of HPAC and related techniques, with an emphasis on their use for the analysis of biological compounds and pharmaceutical agents. Various types of binding agents for these methods will be considered, including antibodies, immunoglobulin-binding proteins, aptamers, enzymes, lectins, transport proteins, lipids, and carbohydrates. Formats that will be discussed for these methods will range from the direct detection of an analyte to indirect detection based on chromatographic immunoassays, as well as schemes based on analyte extraction or depletion, post-column detection, and multi-column systems. The use of biological agents in HPLC for chiral separations will also be considered, along with the use of HPAC as a tool to screen or study biological interactions. Various examples will be presented to illustrate these approaches and their applications in fields such as biochemistry, clinical chemistry, and pharmaceutical research.
1 Introduction
HPLC is an important method for the separation, analysis and characterization of biological, clinical and pharmaceutical samples.1–4 The combination of biologically-related binding agents, or “affinity ligands”, with HPLC has been of great interest in recent years for the creation of selective separation methods for the analysis of targets such as drugs, disease biomarkers, and biopharmaceuticals.1,2,4–6 This has given rise to a technique known as high performance affinity chromatography (HPAC).1–3,5,6 This review will discuss the general principles of HPAC and related techniques, with an emphasis on their use for the analysis of biological compounds and pharmaceutical agents. Various types of binding agents for these methods will be considered, including antibodies, antibody mimics, immunoglobulin-binding proteins, carbohydrates, carbohydrate-binding agents, enzymes, transport proteins, and lipid-based agents(see Table 1).
Table 1.
Examples of biological binding agents that have been used in high performance affinity chromatography and related methods
| Type of binding agent | Applications |
|---|---|
| Antibodies | Sample purification Chromatographic immunoassays Immunoextraction and immunodepletion Post-column immunodetection |
| Immunoglobulin-binding proteins | Antibody measurement of characterization Secondary binding agents for antibody immobilization |
| Aptamers | Small targets separation (nucleotides) Biomacromolecule analysis (thrombin, cyctochrome C, lysozymes). |
| Enzymes | Inhibitor isolation Chiral separations Immobilized enzyme reactors Screening of enzyme targets and inhibitors |
| Lectins | Glycoproteomic anlaysis Glycomic analysis |
| Transport proteins | Chiral separations Solute-protein interaction studies Free drug fraction analysis |
| Lipids | Drug lipophilicity measurements Ligand screening for membrane receptors |
| Carbohydrates | Chiral separations |
2 General principles of affinity-based methods in HPLC
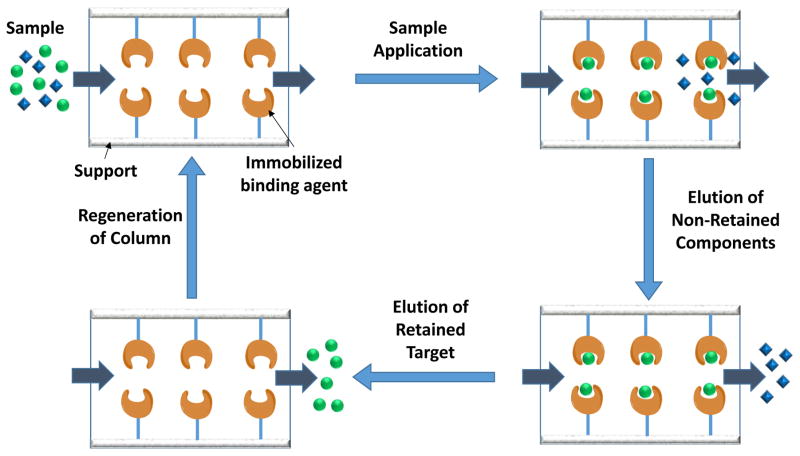
Affinity chromatography is a type of liquid chromatography in which an immobilized biologically-related binding agent, or “affinity ligand”, is used as the stationary phase( see Figure 1).7 The merits of this method include its high selectivity and ability to provide relatively simple separations for specific target compounds, even when this approach is used with complex mixtures such as cell cultures, food samples, and clinical specimens like serum or urine. 1–7 One limitation or requirement for this method is the need for an appropriate binding agent for the given target.4–7 Another requirement is the need for adequate techniques for immobilization of the binding agent and conditions that can be used with this agent for the application and elution of the target for either its purification or analysis.7 The following sections of this review will discuss the various categories of binding agents that have been employed in this method, the types of compounds that bind to these agents, and the conditions under which these binding agents have been used in chromatographic systems for target isolation, analysis or characterization.
Figure 1.
Steps involved in the on/off elution mode of affinity chromatography. The analyte, or target, that can be retained by the immobilized binding agent is represented by the circles, while non-retained sample components are represented by the diamonds.
Traditional affinity chromatography is commonly used for biomolecule purification and generally employs low-performance supports such as agarose or carbohydrate-based materials.1,7,8 HPAC is a type of affinity chromatography that instead uses supports that are suitable for work in HPLC, such as narrow-diameter silica or glass particles, perfusion-based materials, and monolithic supports.4,7,8 This combination provides HPAC with greater speed, precision and ease of automation than traditional affinity chromatography, making HPAC more suitable for use in analytical applications.1,5,8,9 The retention of a solute or analyte in either traditional affinity chromatography and HPAC is based on the specific and reversible interactions that occur between many biological binding agents and their targets.8,9 An example would be the use of an immobilized antibody within a column to bind and capture its given target, or antigen, from applied samples.1,5,7,8,10
There are various ways that affinity ligands can be used in HPAC and related methods. One common format is the on/off elution mode that is shown in Figure 1. In this format, a sample containing the target analyte is applied to an affinity column in the presence of an application buffer.11–13 During this application step, non-retained components will pass through the column, while the analyte is retained by the immobilized binding agent. The analyte is then later eluted by applying an elution buffer, followed by column regeneration.11,12 This format is often used for binding agents that have strong retention for their targets (i.e., association equilibrium constants of roughly 106 M−1 or higher).14 The presence of this strong binding means that an elution buffer that involves a change in the pH, ionic strength, polarity or mobile phase content (e.g., the use of chaotropic agents) is needed to weaken the interaction of the target with the column and to promote the target’s elution.15
It is sometimes possible to use affinity ligands in HPAC under isocratic conditions. The resulting method is known as weak affinity chromatography.16 This situation allows the same mobile phase to be used for both sample application and elution. This approach can be practical when the association equilibrium constant for the target with the immobilized binding agent is less than 105–106 M−1.16 This approach is typically used with affinity columns that are employed in chiral separations17,18 or in studies of biological interactions that have weak-to-moderate binding strengths.1,2,5
3 Affinity methods in HPLC using antibodies
Antibodies, or immunoglubulins, have been used in a variety of ways in HPAC and related affinity methods. The use of antibodies or related agents as stationary phases in chromatography is often referred to as immunoaffinity chromatography (IAC), and the use of these stationary phases in HPLC is known as high performance immunoaffinity chromatography (HPIAC).13,14,19 Antibodies are glycoproteins that are produced by the body in response to a foreign agent, or antigen (e.g., bacteria or a virus).13,14,19 The structure of a typical antibody, using immunoglobulin G (IgG) as an example, consists of a “Y”-shaped structure (e.g., see Figure 2).13,14,19 The lower stem of such an antibody is known as the Fc region and is highly conserved from one type of antibody to the next.13 The upper arms contain two identical antigen-binding sites and are known as the Fab regions; the amino acid sequences of these two regions can be highly variable between different antibodies.13,19 It is this variability that allows the body to produce antibodies against a wide range of targets and to produce antibodies that have strong and selective binding to these targets, with association equilibrium constants that are often in the range of 108–1012M −1.19
Figure 2.
General schemes for (a) a chromatographic displacement immunoassay and (b) a chromatographic sandwich immunoassay.
One format in which HPIAC, IAC and related methods can be used is for the purification or direct detection of an analyte. This type of work is usually done by utilizing the on/off format that was illustrated in Figure 1.11–13 Purification of many pharmaceutical agents and biomolecules have been performed by using IAC with the on/off elution mode, including proteins, glycoproteins, drugs, and environmental agents.1,13,14 A wide variety of analytes have also been analyzed by IAC with direct detection, such as antibodies, hormones, fibrinogen, albumin, β2-microglobulin, transferrin, cytokines, cell receptors and carbohydrates.20 This direct detection approach has further been used to investigate the thermodynamics and kinetics of binding by immobilized antibodies against targets such as the herbicide 2,4-dichlorophenoxyacetic acid and the hormone L-thyroxine.11,12 In addition, antibodies have been used in HPLC columns as chiral stationary phases and with direct detection for such analytes as R/S-abscisic acid, various D/L-amino acids, (R, R)- or (S, S)-finrozole, D/L–hydroxy acids, D/L-lactic acid, S-methamphetamine, and L-triiodothyronine.17,18,21,22
Indirect analysis can also been achieved by using antibodies in HPAC or related methods. The resulting combination is often known as a chromatographic immunoassay or flow-based immunoassay.1,13,14,19 As is demonstrated in Figure 2, these techniques may involve the use of either a competitive binding format (e.g., the simultaneous injection, sequential injection or displacement methods) or a non-competitive format (e.g., a sandwich immunoassay or one-site immunometric assay). In addition, these methods can make use of labels for detection that range from enzymes or fluorescent tags to chemiluminescent agents, liposomes, and radioisotopes.23
A chromatographic competitive binding immunoassay usually involves competition between a target analyte and a labeled analog for immobilized antibodies in a column.13 There are several types of methods that fall in this category.23,24 One of these is the simultaneous injection format, in which a mixture of the target and a fixed amount of a labeled analog of this target are injected together onto a column that contains a small amount of antibodies for the target. Due to competition between the labeled analog and the target for the immobilized antibodies, the amount of labeled analog that is non-retained will increase as the concentration of target in the sample is increased.25 This approach has been employed with fluorescent, chemiluminescent, thermometric and electrochemical detection; examples include the use of labels based on fluorescein, Lucifer yellow, Texas red, Cy5 and acridinium ester or the enzymes horseradish peroxidase (HRP), alkaline phosphatase and catalase.23 Some analytes that have been measured by this format are drugs and hormones like testosterone, thyroxine, cortisol, theophylline, digitoxin and gentamicin or peptides and proteins such as insulin, human serum albumin (HSA), transferrin, and IgG.23 This method has also been employed with the targets carbaryl, isoproturon, and atrazine.23
The sequential injection format is another type of chromatographic competitive binding immunoassay. This method has the target and sample being applied first to the immobilized antibody column, followed later by injection of the labeled analog.26 This approach again gives a response in which the amount of non-retained label increases as the target concentration increases; however, this method avoids the need for placing any label into the sample and can give lower detection limits than the simultaneous injection method.13,19 The sequential injection format has been used for the analysis of imazethapyr, IgG, digoxin, atrazine, and HSA.19,23 Labels that have been utilized in this technique have included fluorescent tags (e.g., fluorescein), enzymes (e.g., β-galactosidase, alkaline phosphatase, and HRP), and liposomes that incorporate such tags or labels.19,23
A displacement immunoassay, which is shown in Figure 2(a), is another type of chromatographic competitive binding technique.19,23 In this method, the labeled analog is first injected onto an antibody column; this is followed by an injection of the target, which can displace some of the bound labeled analog. The size of the displacement peak for the labeled analog will be related directly to the concentration of target in the sample.13,14 This type of assay has been combined with fluorescent labels for the detection of cocaine, benzoylecgonine, and 2,4-dinitrophenol, and has been used with absorbance detection for the analysis of transferrin and HSA.27–29 In addition, this method has been adapted for use with ultrafast affinity extraction for the analysis of free drug and free hormone fractions.30,31 A related method based on a reverse displacement assay has been described (i.e., using the drug phenytoin as a model analyte), in which an analog of the target was immobilized and used to bind a labeled binding agent such as an antibody or Fab fragment.32 In this latter technique, the target was able to bind to some of the labeled agent and cause it to be displaced from the column, giving a signal that was proportional to the target’s concentration.32
Two types of non-competitive chromatographic immunoassays are the one-site immunometric assay and the two-site immunometric assay. In a one-site immunometric assay, a fixed amount of labeled antibodies or antibody fragments is mixed with the sample and target. This mixture is then injected onto a column that contains an immobilized analog of the analyte, which is used to remove any excess labeled antibodies or antibody fragments.13,33 Either the amount of the non-retained or retained labeled agent can then be used to determine the concentration of target that was present in the original sample.1,13 This technique has been used to measure granulocyte colony-stimulating factor, interleukin-10, digoxigenin, α-(difluoromethyl)ornithine, fatty acid binding protein, 17-estradiol, α-fetoprotein, digoxin, and thyroxine.23 These assays have used labels based on fluorescein, HRP, alkaline phosphatase, β-galactosidase, and liposomes containing fluorescent agents.23
The two-site immunometric assay, or sandwich immunoassay, utilizes two different antibodies against the same target, with one type of antibody being immobilized for target extraction and the other being labeled for detection.1,13 The general scheme for this type of assay is shown in Figure 2(b). The amount of target in a sample can be determined by looking at the amount of labeled antibodies that are captured along with the target by the column.1,13 Examples of targets that have been examined by using a chromatographic sandwich immunoassay have included thyroid-stimulating hormone, parathyroid hormone, HSA, IgG, human chorionic gonadotropin, interleukin-5, and anti-IgG antibodies.34–37 These methods have employed labels that make use of absorbance, fluorescence, chemiluminescence orelectrochemical detection.34–37
Immunoextraction is another common application for HPIAC or IAC in pharmaceutical and clinical analysis. This method uses an immobilized antibody column to isolate one or more targets prior to their analysis by a second method. Immunoextraction has been coupled off-line with liquid chromatography (LC), gas chromatography (GC) and capillary electrophoresis (CE) for the analysis of atrazine, bovine serum albumin (BSA), cortisol, clenbuterol and phenytoin in urine, food and water.14,19,38 For instance, an off-line immunoextraction column has been used to bind testosterone and epitestosterone from urine, followed by analysis of the extracted fraction by micellar electrokinetic chromatography.38 On-line immunoextraction has also been coupled with LC, CE and mass spectrometry (MS) for the analysis of drugs, proteins and other targets.19,33,38–40 As an example, one recent study used on-line immunoextraction with HPAC to capture normal or modified forms of HSA and to examine the binding by various drugs with this captured protein.41
Immunodepletion is a method related to immunoextraction that instead uses antibody columns to remove a possible interfering component from a sample.19 This method has often been used in proteomics and the discovery of low-abundance proteins as biomarkers.1,19,42 In this application high abundance proteins (e.g., HSA and IgG in human serum) can be removed by a column that contains antibodies against these proteins. The use of immunodepletion in this manner can make it easier to study and examine low abundance proteins and to analyze the simplified sample by a second method, such as two-dimensional electrophoresis or reversed-phase chromatography.42–45
Post-column immunodetection is an approach in which columns containing antibodies or related binding agents are utilized to monitor specific solutes that are eluting from an HPLC column.20,46 This technique typically uses a post-column reactor to combine and mix the separated components with a labeled agent, which is used for indirect target detection in a chromatographic immunoassay. The post-column reactor is followed by a column that contains an immobilized antibody or antigen that is used to measure an eluting target through the effect this target has on the ability of the labeled agent to bind to this second column.20,46 Post-column immunodetection has been performed with sandwich immunoassays and one-site immunometric assays.14 For instance, a sandwich immunoassay format has been used in a post-column scheme to measuring growth-hormone-releasing factor as it eluted from a reversed-phase column.47 One-site immunometric assays have been employed in post-column methods to monitor digoxin, digoxigenin and human methionyl granulocyte colony-stimulating factor.14 The use of an antibody column for direct detection in a post-column system is also possible if the eluting target is capable of generating enough signal for detection. This last approach has been used to monitor acetylcholinesterase (AChE) that eluted from a size-exclusion column by capturing this target with an anti-AChE antibody column and detecting the AChE by adding a coloring-forming substrate for this enzyme.47
4 Immunoglobulin-binding proteins
Immunoglobulin-binding proteins are bacterial proteins that can be used to bind, purify or capture immunoglobulins from various samples and sources.48 A common example is protein A from Staphylococcus aureus, which is often used to isolate and bind many subclasses of IgG antibodies49 and a number of other immunoglobulin classes (e.g., IgA and IgM from some species).48 Another example is protein G, which is from group G streptococci and which can bind IgG-class antibodies from a wide variety of species.50,51 Protein L, which is obtained from Peptostreptococcus magnus, is another immunoglobulin-binding protein and has strong binding with human IgG, IgA, IgM, and IgE class antibodies.52 In addition, recombinant hybrid proteins such as protein A/G and protein G/L have been developed to expand the types of immunoglobulins that can bind to a single agent in this group.48
Immunoglobulin-binding proteins have been used not only for isolating various types of immunoglobulins and antibodies but also for detecting and measuring these targets in various samples.53–58 This latter application has been of particular interest in recent years with the rapid growth in the market for monoclonal antibodies as biopharmaceutical products.59,60 For instance, protein A has been used in affinity chromatography for the measurement of both monoclonal antibodies and polyclonal antibodies.53,54 Protein G affinity chromatography has also been used to measure monoclonal and polyclonal antibodies.55,58,61 Such applications have involved both particulate supports and perfusion media or monoliths.54–56
A number of reports have used columns containing immunoglobulin-binding proteins in combination with other types of columns. For instance, protein A has been combined with size exclusion chromatography for the simultaneous analysis of host cell proteins, monoclonal antibodies and their aggregates (see Figure 3).62–65 In this approach, a size separation of the sample components that are not retained by a protein A column (e.g., feed impurities or host cell proteins) can be used to provide information that can aid in the consistent production of monoclonal antibodies,66 while a size separation of the retained components can be used to separate and measure antibodies versus antibody aggregates.67
Figure 3.
Chromatogram obtained by combining protein A and size-exclusion HPLC columns for the analysis of monoclonal antibodies versus other components in a Chinese hamster ovary (CHO) cell culture. The peaks for the monoclonal antibodies and their aggregates are shown to the right, after their elution from the protein A column and loading onto the size-exclusion column, and the results for the other sample components (e.g., DNA fragments, host cell proteins, amino acids, nucleotides, etc.) that were non-retained by protein A column are shown to the left, after the first application of the loading buffer and passage of these components through the size-exclusion column. Adapted with permission from Ref. 64.
Protein G has been used in affinity columns in a sandwich immunoassay to detect antibodies in human serum.57 This method has been applied to the analysis of anti-human growth hormone antibodies, anti-BSA antibodies and human IgG.57,68,69 Protein G-based immunoassays have also been carried out in other formats. One example is a dual column immunoassay that combined a protein G column with a reverse-phase column to measure both the titer of antibodies against a specific antigen and the amount of the antigen in injected samples.61
Immunoglobulin-binding proteins can be further employed as secondary binding agents to capture and immobilize antibodies in columns. This approach has been used with protein G columns to immobilize antibodies that could then be employed to bind and measure fluoroquinolones or insulin in serum.70,71 Protein G columns have also been used to adsorb antibodies for use in competitive binding immunoassays.72,73 This latter work has utilized such columns in a simultaneous injection competitive immunoassay to measure HSA72 and in sequential injection-based methods to analyze HSA or transferrin.72,73
5 Aptamers
Aptamers are single strands of DNA or RNA with three-dimensional structures that are capable of binding to specific target compounds.74–77 These binding agents can have relatively large association equilibrium constants, with values that may be as high as 106 to 109 M−1.74,76 The degree and specificity of this binding has made aptamers of interest as antibody mimics for both small and large targets (e.g., proteins, low mass biomolecules, and drugs).74,78 Aptamers have been used within biosensors and/or in columns for the detection of biomarkers or for the purification of various compounds of interest in pharmaceutical and clinical applications. 74,76,77,79
Aptamers can be generated by using a technique known as the systematic evolution of ligands by exponential enrichment, or SELEX.79 SELEX is a multi-step cycling technique in which a large pool of oligonucleotides is screened to eventually acquire sequences which bind to the desired target.80 These sequences are amplified by using the polymerase chain reaction and the screening process is repeated until an aptamer is obtained with both the desired specificity and binding strength for the target.80
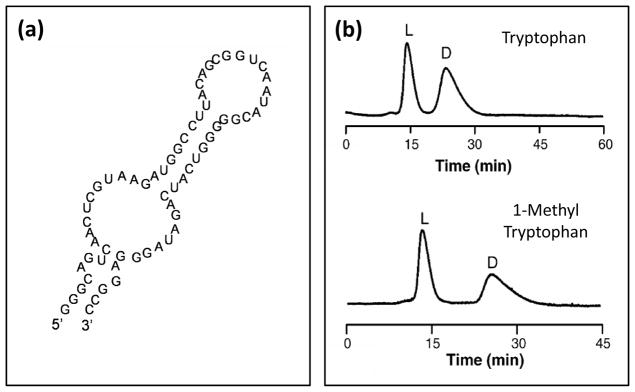
A few studies have examined the use of aptamers in columns that can bind to small targets. Early work in this area explored the use of immobilized aptamers in capillary columns for the separation of adenosine and other nucleotides.81,82 These columns were also used for the analysis of adenosine in vivo in rat brains, and demonstrated good reproducibility, stability and efficiency in such an application.82 In addition, aptamers have been utilized with a perfusion support or silica particles and used as chiral stationary phases for the separation of D- and L-arginine-vasopressin, D- and L-adenosine, and the enantiomers of tyrosine and various related analogs (see Figure 4).83–86
Figure 4.
(a) Sequence of an L-RNA aptamer that was used as a binding agent for L-tyrosine and related compounds and (b) chromatograms that were obtained when using this aptamer in an HPLC column for the chiral separation of D/L-tryptophan (top) and D/L-1-methyl tryptophan (bottom). Adapted with permission from Ref. 86.
Aptamers have also been employed in columns for the separation and analysis of biomacromolecules. For example, a biotinylated DNA aptamer has been bound to immobilized streptavidin on beaded polyacrylamide and used to purify a recombinant human L-selectin–receptor globulin fusion protein from a Chinese hamster ovary cell culture.87 Two aptamer sequences were developed as affinity ligands for thrombin and used in a chromatographic sandwich assay for this target, giving a detection limit of 0.1 nM without the need for labeling or derivatization.88 A biotinylated aptamer against cyctochrome C was bound to a glycidyl methacrylate/trimethylolpropane trimethacrylate (GMA/TRIM) monolith containing immobilized streptavidin and used to separate cytochrome C and thrombin from a mixture of several proteins; this technique was then used to detect cytochrome C in a liver tissue lysate and thrombin in serum and blood.89 Another report used an aptamer that was covalently immobilized in a GMA/ethylene dimethacrylate (GMA/EDMA) monolith for the extraction and screening of lysozymes in samples such as chicken egg white.90 In addition, an organic-inorganic hybrid silica monolith was used to immobilize an aptamer for the binding and detection of human α-thrombin.91
Aptamer affinity chromatography has been used in several studies with capillary columns81,82,88,89,91,92 and microfluidic devices.93,94 For instance, aptamers that could bind to thrombin were covalently attached to the inner surface of a bare fused silica capillary and used to extract thrombin from a protein mixture, with the thrombin being detected and identified by fluorescent labeling and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS).92 A microfluidic device has been used with aptamer affinity chromatography for the thermally-controlled extraction and elution of adenosine monophosphate.93 Another group developed a microfluidic device capable of photolytic elution by using a photo-cleavable linker to immobilize an RNA aptamer.94 This RNA aptamer was used to retain several proteins, which were then released and eluted after exposure of the aptamer to UV light.94
6 Enzymes
There have been a number of applications for affinity chromatography involving work with enzymes as either targets or immobilized binding agents. For instance, the use of enzymes in therapeutics has created a demand for improved methods for separating and purifying these agents.95–97 One advantage of using enzymes as therapeutics is their ability to often bind their substrate or target with great affinity and selectivity.95,97 Another advantage of enzymes is their capability to catalytically convert their substrate into a product in an efficient manner.95,97 Affinity separations for enzymes have often used immobilized substrates, inhibitors or cofactors as the binding agents.1 Specific examples are the use of immobilized flavin nucleotides for the extraction of flavin adenine dinucleotide synthetase from rat liver98 and the use of furin-specific inhibitors for purification of the proprotein/prohormone convertase furin from ovarian cells.99
There have been a few cases in which immobilized enzymes have been used directly for the isolation or extraction of targets. An example is work in which carboxypeptidase A was immobilized and used for the isolation of carboxypeptidase inhibitor.100 Another study used immobilized pepsin to isolate major and minor forms of a pepsin inhibitor from a parasite.101 The immobilization of enzymes in columns can often be done in a manner that does not drastically affect the activity or selectivity of the enzyme.96,102–104 An additional advantage of using immobilized enzymes is their good stability and ability to be reused multiple times, while providing a format that allows easy removal of the bound targets or products from the enzyme.102,104
Another application of immobilized enzymes has been their utilization as chiral stationary phases.105–114 This approach makes use of the ability of some enzymes to select between the enantiomers of solutes that interact with their binding sites.106 For example, immobilized penicillin G acylase has been employed in chiral separations for several acidic aryl agents and non-steroidal anti-inflammatory drugs,107–109 while glucoamylase G1 and G2 have been used as chiral stationary phases for amino alcohols.110,111 Other enzymes that have been used as chiral selectors are cellobiohydrolase I, lysozyme, trypsin, α-chymotrypsin, and pepsin.105,112–114
A related field to affinity chromatography is that of immobilized enzyme reactors, or IMERs.115–119 Analytical applications of IMERs have included their used for screening potential enzyme inhibitors, examining drug metabolism pathways, and digesting proteins for peptide mapping.1,115–119 IMERs for chromatographic systems have been made using silica, agarose and various types of monoliths as the support.120–124 In addition, IMERs have been used in such formats as HPLC, CE and microfluidics.1,115–119,121,125–127
7 Lectins
Lectins are another group of binding agents that have been used in affinity chromatography and HPAC. Lectins are non-immune system proteins that can bind reversibly to specific carbohydrate groups.128–131 This property has made lectins important as tools for capturing and studying glycoconjugates such as glycoproteins.129 One lectin that has often been used in affinity columns is concanavalin A ( Con A) from Canavalia ensiformis, which can bind to high-mannose type glycans, hybrid type glycans with mannose branching, and complex type biantennary glycans.132–134 Another lectin that has frequently been used as an affinity ligand is wheat germ agglutinin (WGA) from Triticum vulgare, which binds to sialic acid and the core portion of N-glycans, such as N, N-diacetylchitobiose (also known as (GlcNAc)2), and the tetrasaccharide GlcNAcβ1–4Manβ1–4GlcNAcβ1–4GlcNAc which is present in bisected N-glycans.135 Other examples of lectins that have been employed in affinity methods are Aleuria aurantia lectin (AAL) and Sambucus nigra agglutinin (SNA), which bind to α1–6, α1–2, α1–3 linked fucoses and α2–6 linked sialic acids.136,137 Artocarpus lectin (AIL or jacalin) has also been used and binds Galβ1-3GalNAcor high mannose type glycan s.138,139
The selective enrichment of carbohydrate-containing targets like glycoproteins or glycopeptides by lectins has become important in studying complex biological mixtures.42,138,140,141 For instance, Con A and WGA both bind to asparagine-linked glycans, while jacalin can bind to serine- or threonine-linked glycans.138,140 These binding specificities have been combined in a multi-lectin column where Con A, WGA and jacalin were each immobilized to agarose and used to capture a wide range of glycoproteins from human serum.138 Multi-lectin affinity chromatography was also used with SDS-PAGE and LC-MS/MS to identify biomarkers for diabetic nephropathy in human plasma,141 and multi-lectin columns have been utilized in the analysis of cellular lysates.142,143
Several reports have combined immunodepletion with lectin columns. For instance, the immunodepletion of twelve high-abundance proteins was used along with a column containing immobilized Con A and WGA and detection based on quadrupole TOF-MS to measure low abundance glycoproteins in human serum.42 An automated platform was developed by coupling an immunodepletion column on-line with a multi-lectin column;144 this platform was used to deplete abundant serum proteins prior to the enrichment and analysis of lower-abundance glycoproteins.145,146 The fractions collected from this multi-dimensional system have been subjected to isoelectric focusing, followed by with-in gel digestion and LC-MS analysis.147
When several lectin columns are coupled together, the resulting technique is known as serial lectin affinity chromatography (SLAC).139 For instance, high-mannose N-linked glycan-containing glycoproteins have been depleted by using a Con A column, followed by use of a jacalin column to capture O-linked glycan-containing glycoproteins.139 Serial lectin affinity chromatography has been used to characterize co-existing glycans on the same glycoconjugates by altering the order.148 SLAC can also be used to assess the degree of a specific type of glycosylation. For instance, SLAC with Con A and SNA has been used in parallel with Con A affinity chromatography and stable isotope coding to compare the degree of α2–6 sialylation on hybrid, high-mannose and complex biantennary glycans.149 In another example, sialic acid-bearing glycoforms with complex-type glycans were fractionated into bi-antennary and tri-, tetra-antennary glycoforms by utilizingserial SNA/Con A affinity chromatography. 150
Lectins have been employed with various high performance materials and column formats. One report used a polystyrene-divinylbenzene perfusion support with immobilized Con A, WGA and jacalin to make a multi-lectin column.151 This column was combined with SDS-PAGE, LC-MS, isoelectric focusing and an antibody microarray to identify breast cancer-related glycoproteins in human blood.152 A silica-based microcolumn was used to immobilize Con A SNA, Ulex europaeus lectin (UEA-I) and Phaseolus vulgaris agglutinin (PHA-L), enabling work with microscale samples of human serum.137,153,154 Macroporous silica particles with a diameter of 1.6 μm were used with Con A and AAL to enrich glycoproteins from human serum and to perform glycomic and glycoproteomic studies.155
Several monolith supports have also been employed with lectins. One study compared the use of immobilized Con A and WGA on a cationic monolith based on 2-(methacryloyloxy)ethyl]trimethyl ammonium chloride (MAETA) or based on a neutral GMA/EDMA monolith, with the cationic monolith allowing the concentration and detection of glycoproteins at levels down to roughly 10−8 M.156 A capillary monolith was developed in which Con A was immobilized onto GMA/EDMA that had been modified with Cu2+-iminodiacetic acid; this type of column doubled the amount of active binding sites and was used to enrich and compare glycoproteins in samples such as urine.157 Another study placed gold nanoparticles onto an EDMA-based monolith, which were then used to immobilize Erythrina cristagalli lectin (ECL) for the enrichment of galactosylated proteins.158 In addition, Con A has been immobilized onto a nanoporous gold monolith and used to capture ovalbumin from a mixture of this protein and BSA.159
8 Transport proteins
Serum transport proteins and related agents have been used for a number of applications in affinity chromatography and HPAC.2 One example of this type of agent is HSA. HSA is the most abundant protein in human plasma, with a normal concentration of 35–50 g/L. HSA and the closely-related protein BSA can interact with various solutes that include many drugs, fatty acid s, and low mass hormones.160,161 The two major binding sites on HSA for drugs are often referred to as Sudlow sites I and II.162 A second example of a serum transport protein is alpha1-acid glycoprotein (AGP). AGP is an acute phase glycoprotein that has a normal plasma concentration of 0.5–1.0 g/Land a carbohydrate content as high as 45%( w/w).163–165 AGP is an acidic protein, with an isoelectric point of 2.8–3.8, and can bind to a number of basic, neutral or cationic drugs.2,166 Other examples of serum binding agents for small solutes are low density lipoprotein (LDL) and high density lipoprotein (HDL).167–175 These lipoproteins are complex particles of proteins and lipids that transport substances such as triglycerides and cholesterol esters,172–175 as well as some drugs.167–175
Serum proteins have been widely used as chiral stationary phases.166 For instance, HSA and BSA-based columns have been used to separate many acidic and neutral chiral compounds that have included coumarins (e.g., warfarin), benzodiazepine derivatives, barbiturates, benzothiadiazeines, α-arylpropionic acids (e.g., ibuprofen), and some amino acids (e.g., tryptophan).176–181 Factors that can be varied to optimize these separations are the pH of the mobile phase166,179,182, the use of organic modifiers in the mobile phase (e.g., 1-propanol) and the ionic strength of the mobile phase166,183 Many chiral HPLC methods have used these binding agents with silica particles,176–181 although some work has also employed silica monoliths or organic monoliths.183–186
AGP has also been used as a chiral stationary phase for a variety of compounds.187–194 A few examples are the chiral separation and analysis of atenolol, citalopram, warfarin, verapamil, norverapamil, atropine, and vinca alkaloid analogs.187–191 Liquid chromatography-tandem mass spectrometry has been used with AGP columns to separate and quantitate racemic drugs such as methadone, ornidazole, azelnidipine, ondansetron, and doxazosin.192–196 These efforts have used AGP with both silica particles and silica monoliths.197,198 Factors that can be used to adjust these separations have again included temperature and the pH or composition of the mobile phase (e.g., the use of organic modifiers such as methanol, ethanol, propanol or a tertiary alcohol).199,200
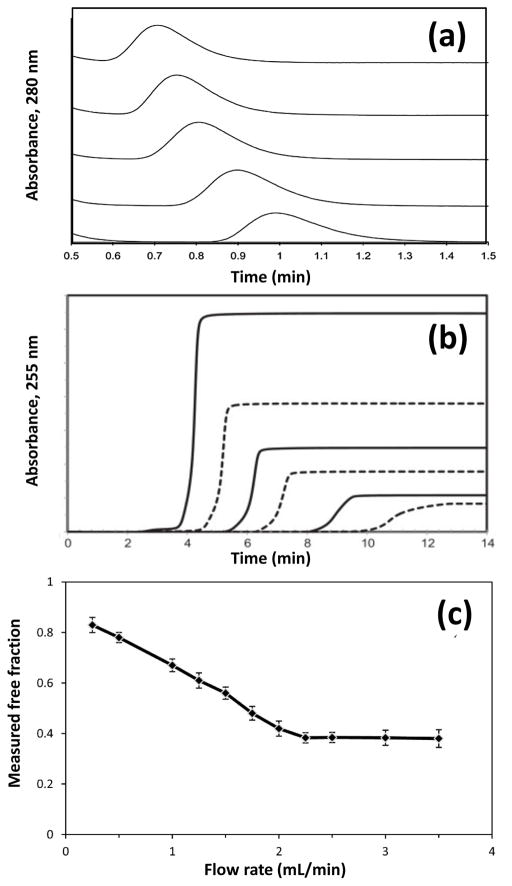
Several types of columns containing serum proteins have been used as tools to study solute-protein interactions.2 One way this can be done is through zonal elution experiments, as shown in Figure 5(a). In a zonal elution experiment, a narrow plug of an analyte solution is injected onto a column that contains an immobilized agent (e.g., a serum transport protein). The elution of this sample plus is then monitored, with the retention time of the analyte being related to the equilibrium constant for this analyte to the immobilized binding agent. For instance, the retention time for an injected probe compound that has a known binding site on an immobilized protein can be monitored while various concentrations of a possible competing agent are placed into the mobile phase.201 This type of experiment can make it possible to determine how the injected and added solutes compete for a given site on the protein and to measure the affinities of these solutes at this site. For instance, this method has been used to examine site-specific interactions of drugs such as acetohexamide and tolbutamide with HSA202 and to compare the binding sites of carbamazepine and propranolol on AGP.203 Zonal elution can also be employed to examine the effects on this binding when there is a change in the temperature or the solvent pH and composition.204–206
Figure 5.
Examples showing the use of HPAC to study biological interactions based on (a) competition studies in zonal elution experiments, (b) frontal analysis, or (c) ultrafast affinity extraction. The results in (a) are for the injection of L-tryptophan as a site-selective probe onto a 2 cm × 2.1 mm I.D. column containing immobilized HSA columns and in the presence of various concentrations of tolbutamide in the mobile phase (left-to-right: 20, 15, 10, 5, or 1 μM). The chromatograms in (b) were obtained for glimepiride that was applied to a 2 cm × 2.1 mm I.D. column containing HSA; the glimepiride concentrations were 50, 30, 20, 15, 10, or 5 μM (top-to-bottom). The data in (c) shows the effect of the injection flow rate on the measured free fraction for verapamil in a sample that contained a mixture of 10 μM verapamil and 20 μM AGP that was applied to a 0.5 cm × 2.1 mm I.D. AGP microcolumn. These figures are adapted with permission from Refs. 201, 202, and 219.
Frontal analysis is another method that has been used with immobilized serum proteins to investigate solute-protein interactions. In this technique, a known concentration of a solute is continuously applied to a column to titrate binding sites on an immobilized agent. A breakthrough curve is then obtained, as shown in Figure 5(b), in which the mean position of this curve is related to the concentration of the applied solute, the number of binding sites for the solute in the column, and the equilibrium constants for the solute at these sites. This technique has been used to examine the interactions of HSA with R/S-warfarin and D/L-tryptophan (as well as coumarin or indole derivatives), thyroxine, acetohexamide, carbamazepine, imipramine, lidocaine, phenytoin, tolbutamide, and verapamil.180,181,202,207–215 Frontal analysis has also been used to examine the binding of carbamazepine and lidocaine with AGP203,211 and the interactions of R- and S-propranolol with HDL, LDL and very low density lipoprotein.216,217 This method can also be employed to examine the effects of temperature and solvent competition on these interactions, as well as solute-solute competition or displacement.204,205
Another way serum proteins have been used in HPAC is in the method of ultrafast affinity extraction, as is illustrated in Figure 5(c).218,219 In this technique, a sample of the solute is prepared in the presence or absence of a soluble binding agent such as a serum protein. This sample is then applied onto an affinity microcolumn for extraction of the solute, which is often done at a high flow rate.218 One way this method can be used is to capture the non-protein bound (or free) form of the solute as the sample passes through the column.220–222 This method has been used to measure free drug and hormone fractions in clinical samples and to measure the equilibrium and rate constants for drug or hormone interactions with proteins in solution.30–32,220–227 Affinity ligands that have been used in microcolumns for this purpose have included both antibodies and serum proteins such as HSA or AGP.30–32,220–227 Ultrafast affinity extraction has also been employed in multi-dimensional HPAC to study drug interactions in clinical samples222,227 and to examine protein interactions with multiple solutes (e.g., chiral drug mixture, such as R/S -warfarin)223 or for solutes that may have multiple binding agentsin a sample.226
9 Lipids
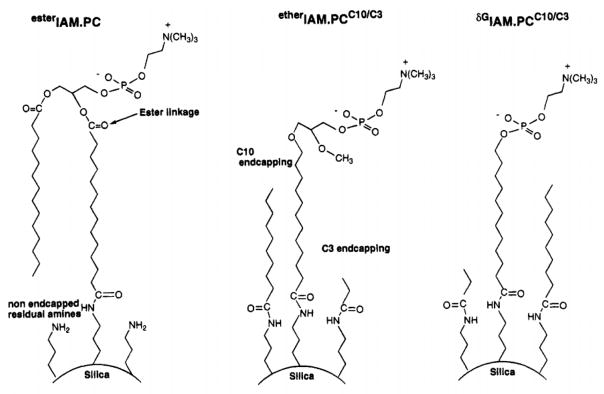
Lipids have explored for use in some applications of HPAC and affinity-based separations. One reason this area has been of interest is because drug lipophilicity directly affects the degree to which pharmaceutical agents can undergo partitioning into cell membranes and passive membrane transport.228 Immobilized artificial membrane (IAM) chromatography is a technique that been employed with lipids to estimate the partition coefficients of drugs for cell membranes229 and to study the oral absorption, blood brain barrier permeability, skin permeability, and pharmacokinetics of various drugs.230 The artificial membranes that are used in these columns consist of monolayers of phospholipid analogs that are covalently attached to a support (see Figure 6).231,232 Phosphatidylcholine, which is a major component of biological membranes, is the most common agent that has been immobilized for this purpose.233 Supports containing sphingomyelin and cholesterol have also been recently created and tested for use as stationary phases in IAM chromatography.234
Figure 6.
Examples of three stationary phases used in immobilized artificial membrane (IAM) columns based on phosphatidylcholine (PC). Reproduced with permission from Ref. 232.
Another application for the columns in IAM has been as platforms for the immobilization of receptors or transporters that are normally found in cell membranes.235–239 The resulting columns have been used to screen the binding of drug candidates to these receptors and transporters.235 These columns can also be used with techniques such as frontal analysis and zonal elution, as described in Section 8, to determine the equilibrium constant sand rate constants for these interactions. For instance, nicotinic acetylcholine receptors have been placed on IAM supports and used with frontal analysis to determine the binding constants of these receptors for (±)-epibatidine, 3-(2(s)-azetidinylmethoxy) pyridine, (−)-nicotine, carbachol and atropine.236,237 P-Glycoprotein has been immobilized onto IAM supports to examine the allosteric interactions and binding constants of this binding agent with doxorubicin, vinblastine, cyclosporin A and verapamil.238,239
Immobilized liposome chromatography (ILC) is a related technique that has been employed in characterizing drug-membrane interactions.240 In ILC, liposomes or lipid bilayers are immobilized onto a support by methods such as entrapment, the binding by these agents to immobilized hydrophobic ligands, or the adsorption of a biotinylated from of such an agent onto immobilized avidin.241–243 Several studies have used ILC to screen the bioactive ingredients or membrane penetrable components in traditional Chinese medicine244–249 and to estimate the affinities of these interactions245,247,248
Monoliths have been used with immobilized lipids, as has been demonstrated with a cholesterol-containing polymeric support.250 This stationary phase has been used with compounds such as alkylbenzenes, steroid hormones and polycyclic aromatic hydrocarbons, giving retention properties that are distinct from both an IAM stationary phase and stationary phases that are commonly used in reversed phase chromatography.251,252 A method for immobilizing phosphatidylcholine in a GMA/EDMA monolith has also been described253,254 and applied to the study of drug-membrane interactions.254 In addition, liposomes have been immobilized within a silica monolith and used to test the interactions of these liposomes with a variety of neutral, acidic or basic drugs. 246
10 Carbohydrates
A variety of carbohydrate-based agents have been used in HPAC and affinity chromatography for pharmaceutical and biomedical analysis. One important application of these ligands has been their use in the separation and analysis of chiral drugs.255 Polysaccharides such as cellulose and amylose are commonly used in chiral stationary phases for HPLC and many such columns are commercially available.256,257,258 Examples are 3,5-dimethylphenyl carbamate phases that make use of cellulose or amylose, as well as 4-methyl benzoate-based chiral stationary phases.256
Cyclodextrinsare another group of carbohydrate-based agents that have been widely used as chiral stationary phases. Cyclodextrins have a torus-like arrangement that contain six, seven or eight glucopyranose units, giving agents known as α-cyclodextrin, β-cyclodextrin, or γ-cyclodextrin, respectively.259,260 The chiral recognition of a solute by a cyclodextrin is related to the relatively hydrophobic cavity and a hydrophilic exterior of the cyclodextrin, which contains a specific arrangement of alcohol groups about the mouth of the cavity. The process of chiral selection involves formation of inclusion complex between a solute and cavity of the cyclodextrin, along with differential binding of the analyte to 2- and 3-hydroxyl groups at the mouth of the cavity.255,261
Cyclodextrin columns have been used in various ways for chiral separations.262–278 This has included work with both reversed-phase and HILIC type mobile phases.262,263 A combination of the reversed-phase and HILIC modes has been used on a β-cyclodextrin stationary phase for the analysis of traditional Chinese medicine.263 In addition, a restricted access β-cyclodextrin column has been used for the direct injection and analysis of chlorthalidone, aminoglutethimide, amlodipine and chlorpheniramine in samples containing BSA.264 Cationic cyclodextrin derivatives have been immobilized to silica for use in the separation of acidic enantiomers265–267 such as dansyl amino acid, carboxylic aryl compounds and flavonoids.266 Phenylcarbamoylated cyclodextrin derivatives that can undergo π-π and dipole-dipole interactions with chiral analytes have been developed as well;268 these agents were used in a bilayer stationary phase that contained both non-derivatized and phenylcarbamoylated-β-cyclodextrin to separate isoxazolines, flavonoids and dansyl amino acids.269 A number of recent studies have examined the use of ‘click’ chemistry for the immobilization of cyclodextrins onto chromatographic supports.270–273 The use of both silica monoliths and organic monoliths have also been considered for use with immobilized cyclodextrins in chiral separations.274–278
Aliphatic and aromatic functionalized cyclofructans (CFs) are another set of carbohydrate-related agents that have been examined for use in chiral separations.279 Cyclofructans are macrocyclic oligosaccharides that contain six or more molecules of D-fructofuranose that are coupled through a β2-1 linkage. Cyclofrutans can be classified based on the number of fructose units they contain, giving compounds such as cyclofructan 6 (CF6), cyclofructan 7 (CF7), and cyclofructan 8 (CF8).279 Nearly 120 racemic primary amine-containing compounds have been separated on an isopropyl-carbamate functionalized CF6 stationary phase.280 Stationary phases containing R-naphthylethylcarbamate CF6 or dimethylphenyl-carbamate CF7 were also evaluated for their chiral recognition of a wide range of chiral acids, amines, metal complexes and neutral compounds. A total of 94 chiral compounds were resolved by either column, which accounted for 43% of compounds in a set of randomly-chosen potential analytes.281
11 Conclusion
This review has discussed how various biologically-related binding agents have been employed in HPAC and related methods as tools for a number of pharmaceutical and biomedical applications. It was shown that many binding agents are available for this work, ranging from antibodies and antibody mimics (e.g., aptamers) to enzymes, transport proteins, immunoglobulin-binding proteins, lectins, carbohydrates, and lipids. The specific and reversible interactions of these agents with their targets have made these binding agents useful as stationary phases in various separation formats. One common example is the on/off elution mode, which can be used for direct analyte detection, affinity-based extraction, or the depletion of a given target from a sample (e.g., as occurs in immunodepletion). Various techniques for indirect detection are also possible in HPAC and related methods, as might occur in chromatographic immunoassays that employ competitive binding or non-competitive binding formats. Other uses described for these binding agents in chromatographic systems have included post-column detection, multi-column systems, chiral separations, and the study of biological interactions. These efforts have also explored the use of various supports with these agents, such as particulate supports, monoliths, and mixed-bed materials. As additional types of supports, binding agents, and formats are developed for these binding agents in HPAC, it is expected that an even greater range of applications will appear for these methods in the analysis and characterization of samples in fields such as biochemistry, clinical chemistry and pharmaceutical research.
Acknowledgments
This work was supported by the National Institutes of Health under grants R01 GM044931 and R01 DK069629, and the National Science Foundation (NSF) under grant CHE 1309806.
References
- 1.Hage DS, Anguizola JA, Bi C, Li R, Matsuda R, Papastavros E, Pfaunmiller E, Vargas J, Zheng X. J Pharm Biomed Anal. 2012;69:93–105. doi: 10.1016/j.jpba.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hage DS, Anguizola J, Barnaby O, Jackson A, Yoo MJ, Papastavros E, Pfaunmiller E, Sobansky M, Tong ZH. Curr Drug Metab. 2011;12:313–328. doi: 10.2174/138920011795202938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moser AC, Hage DS. Electrophoresis. 2008;29:3279–3295. doi: 10.1002/elps.200700871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfaunmiller EL, Bas J, Brooks M, Milanuk M, Rodriguez E, Vargas J, Matsuda R, Hage DS. In: Analytical Separation Science. Anderson JL, Berthod A, Pino V, Stalcup AM, editors. WILEY-VCH; Weinheim: 2015. pp. 461–482. [Google Scholar]
- 5.Hage DS, Anguizola JA, Jackson AJ, Matsuda R, Papastavros E, Pfaunmiller E, Tong Z, Vargas-Badilla J, Yoo MJ, Zheng X. Anal Methods. 2011;3:1449. doi: 10.1039/C1AY05068K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Deeb S, Wätzig H, El-Hady DA. Trends Anal Chem. 2013;48:112–131. [Google Scholar]
- 7.Hage DS, Ruhn PF. In: Handbook of Affinity Chromatography. 2. Hage DS, editor. CRC Press; Boca Raton: 2005. pp. 3–13. [Google Scholar]
- 8.Hage DS. In: Encyclopedia of Chromatrography. 3. Cazes J, editor. CRC Press; Boca Raton: 2010. pp. 17–23. [Google Scholar]
- 9.Yoo MJ, Hage DS. In: Monolithic Chromatography and its Modern Applications. Wang PG, editor. ILM; Glendale: 2010. pp. 3–25. [Google Scholar]
- 10.Hage DS, Jackson A, Sobansky MR, Schiel JE, Yoo MJ, Joseph KS. J Sep Sci. 2009;32:835–853. doi: 10.1002/jssc.200800640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson MA, Moser A, Hage DS. J Chromatogr B. 2010;878:165–171. doi: 10.1016/j.jchromb.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfaunmiller E, Moser AC, Hage DS. Methods. 2012;56:130–135. doi: 10.1016/j.ymeth.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfaunmiller EL, Bi B, Li Z, Vargas-Badilla J, Zheng X, Hage DS. In: Advances in Liquid Chromatography: New Developments in Stationary Phases and Supports for Drugs and Bioanalytical Applications. Hage DS, editor. Future Medicine Ltd; London: 2015. pp. 60–74. [Google Scholar]
- 14.Hage DS, Phillips TM. In: Handbook of Affinity Chromatography. 2. Hage DS, editor. CRC Press; Boca Raton: 2005. pp. 127–172. [Google Scholar]
- 15.Walters RR. Anal Chem. 1985;57:1099A–1114A. doi: 10.1021/ac00288a001. [DOI] [PubMed] [Google Scholar]
- 16.Hage DS, Anne Nelson M, Xuan H. In: Handbook of Affinity Chromatography. 2. Hage DS, editor. CRC Press; Boca Raton: 2005. pp. 79–97. [Google Scholar]
- 17.Hofstetter H, Hofstetter O. Trends Anal Chem. 2005;24:869–879. [Google Scholar]
- 18.Koidl J, Hodl H, Schmid MG, Konrad M, Petschauer S, Kostner GM, Gubitz G. J Biochem Biophys Methods. 2006;69:33–42. doi: 10.1016/j.jbbm.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 19.Moser AC, Hage DS. Bioanalysis. 2010;2:769–790. doi: 10.4155/bio.10.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hage DS. J Chromatogr B. 1998;715:3–28. doi: 10.1016/s0378-4347(97)00621-x. [DOI] [PubMed] [Google Scholar]
- 21.Franco EJ, Hofstetter H, Hofstetter O. J Pharm Biomed Anal. 2009;49:1088–1091. doi: 10.1016/j.jpba.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 22.Zeleke JM, Smith GB, Hofstetter H, Hofstetter O. Chirality. 2006;18:544–550. doi: 10.1002/chir.20286. [DOI] [PubMed] [Google Scholar]
- 23.Hage DS, Moser AC. In: Handbook of Affinity Chromatography. 2. Hage DS, editor. CRC Press; Boca Raton: 2005. pp. 789–836. [Google Scholar]
- 24.Matsuda R, Rodriguez E, Suresh D, Hage D. Bioanalysis. 2015;7:2947–2966. doi: 10.4155/bio.15.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hage DS, Thomas DH, Chowdhuri AR, Clarke W. Anal Chem. 1999;71:2965–2975. doi: 10.1021/ac990070s. [DOI] [PubMed] [Google Scholar]
- 26.Hage DS, Thomas DH, Beck MS. Anal Chem. 1993;65:1622–1630. doi: 10.1021/ac00059a023. [DOI] [PubMed] [Google Scholar]
- 27.Hage DS, Taylor B, Kao PC. Clin Chem. 1992;38:1494–1500. [PubMed] [Google Scholar]
- 28.Wemhoff GA, Rabbany SY, Kusterbeck AW, Ogert RA, Bredehorst R, Ligler FS. J Immunoligical Methods. 1992;8:223–230. doi: 10.1016/0022-1759(92)90029-s. [DOI] [PubMed] [Google Scholar]
- 29.Whelan JP, Kusterbeck AW, Wemhoff GA, Bredehorst R, Ligler FS. Anal Chem. 1993;5:3561–3565. [Google Scholar]
- 30.Clarke W, Schiel JE, Moser A, Hage DS. Anal Chem. 2005;77:1859–66. doi: 10.1021/ac040127x. [DOI] [PubMed] [Google Scholar]
- 31.Ohnmacht CM, Schiel JE, Hage DS. Anal Chem. 2006;78:7547–7556. doi: 10.1021/ac061215f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiel JE, Tong Z, Sakulthaew C, Hage DS. Anal Chem. 2011;83:9384–9390. doi: 10.1021/ac201973v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfaunmiller EL, Anguizola JA, Milanuk ML, Papastavros E, Carter N, Matsuda R, Zheng X, Hage DS. J Chromatogr A. 2014;1366:92–100. doi: 10.1016/j.chroma.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karube I, Matsunaga T, Satoh T, Suzuki S. Anal Chim Acta. 1984;156:283–287. [Google Scholar]
- 35.Hage DS, Kao PC. Anal Chem. 1991;63:586–595. doi: 10.1021/ac00006a008. [DOI] [PubMed] [Google Scholar]
- 36.Johns MA, Rosengarten LK, Jackson M, Regnier FE. J Chromatogr A. 1996;743:195–206. doi: 10.1016/0021-9673(96)00370-6. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Wang Y, Luo G, Yeung WSB. J Liq Chromatogr Relat Technol. 2001;24:1953–1963. [Google Scholar]
- 38.Chen HX, Deng QP, Zhang LW, Zhang XX. Talanta. 2009;78:464–470. doi: 10.1016/j.talanta.2008.11.048. [DOI] [PubMed] [Google Scholar]
- 39.Chen HX, Huang T, Zhang XX. Talanta. 2009;78:259–264. doi: 10.1016/j.talanta.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Jiang T, Mallik R, Hage DS. Anal Chem. 2005;77:2362–2372. doi: 10.1021/ac0483668. [DOI] [PubMed] [Google Scholar]
- 41.Matsuda R, Jobe D, Beyersdorf J, Hage DS. J Chromatogr A. 2015;1416:112–120. doi: 10.1016/j.chroma.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao P, Ren Y, Xie Y. J Chromatogr B. 2009;877:1657–1666. doi: 10.1016/j.jchromb.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 43.Chromy BA, Gonzales AD, Perkins J, Choi MW, Corzett MH, Chang BC, Corzett CH, McCutchen-Maloney SL. J Proteome Res. 2004;3:1120–1127. doi: 10.1021/pr049921p. [DOI] [PubMed] [Google Scholar]
- 44.Dowling P, O’Driscoll L, Meleady P, Henry M, Roy S, Ballot J, Moriarty M, Crown J, Clynes M. Electrophoresis. 2007;28:4302–4310. doi: 10.1002/elps.200700246. [DOI] [PubMed] [Google Scholar]
- 45.Cellar NA, Karnoup AS, Albers DR, Langhorst ML, Young SA. J Chromatogr B. 2009;877:79–85. doi: 10.1016/j.jchromb.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 46.Irth H, Oosterkamp AJ, Tjaden UR, van der Greef J. Trends Anal Chem. 1995;14:355–361. [Google Scholar]
- 47.Vanderlaan M, Lotti R, Siek G, King D, Goldstein M. J Chromatogr A. 1995;711:23–31. doi: 10.1016/0021-9673(95)00059-v. [DOI] [PubMed] [Google Scholar]
- 48.Phillips TM. In: Handbook of Affinity Chromatography. 2. Hage DS, editor. CRC Press; Boca Raton: 2005. pp. 367–397. [Google Scholar]
- 49.Kronvall G, Frommel D. Immunochemistry. 1970;7:124–127. doi: 10.1016/0019-2791(70)90036-4. [DOI] [PubMed] [Google Scholar]
- 50.Björck L, Kronvall G. J Immunol. 1984;133:969–974. [PubMed] [Google Scholar]
- 51.Akerström B, Björck L. J Biol Chem. 1986;261:10240–10247. [PubMed] [Google Scholar]
- 52.Akerström B, Björck L. J Biol Chem. 1989;264:19740–19746. [PubMed] [Google Scholar]
- 53.Compton BJ, Lewis M, Whigham F, Gerald JS, Countryman GE. Anal Chem. 1989;61:1314–1317. doi: 10.1021/ac00188a003. [DOI] [PubMed] [Google Scholar]
- 54.Paliwal SK, Nadler TK, Wang DIC, Regnier FE. Anal Chem. 1993;65:3363–3367. doi: 10.1021/ac00071a005. [DOI] [PubMed] [Google Scholar]
- 55.Gadowski L, Abdul-Wajid A. J Chromatogr A. 1995;715:241–245. doi: 10.1016/0021-9673(95)00614-s. [DOI] [PubMed] [Google Scholar]
- 56.Tscheliessnig A, Jungbauer A. J Chromatogr A. 2009;1216:2676–2682. doi: 10.1016/j.chroma.2008.07.063. [DOI] [PubMed] [Google Scholar]
- 57.Riggin A, Regnier FE, Sportsman JR. Anal Chem. 1991;63:468–474. doi: 10.1021/ac00005a017. [DOI] [PubMed] [Google Scholar]
- 58.Blank GS, Vetterlein D. Anal Biochem. 1990;190:317–320. doi: 10.1016/0003-2697(90)90201-j. [DOI] [PubMed] [Google Scholar]
- 59.Ecker DM, Jones SD, Levine HL, Ecker DM, Jones SD, Levine HL. MAbs. 2015;7:9–14. doi: 10.4161/19420862.2015.989042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rathore AS. Trends Biotechnol. 2009;27:546–553. doi: 10.1016/j.tibtech.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 61.Janis LJ, Regnier FE. Anal Chem. 1989;61:1901–1906. doi: 10.1021/ac00192a024. [DOI] [PubMed] [Google Scholar]
- 62.Horak J, Ronacher A, Lindner W. J Chromatogr A. 2010;1217:5092–5102. doi: 10.1016/j.chroma.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 63.Lemmerer M, London AS, Panicucci A, Gutierrez-Vargas C, Lihon M, Dreier P. J Immunol Methods. 2013;393:81–85. doi: 10.1016/j.jim.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 64.Gjoka X, Schofield M, Cvetkovic A, Gantier R. J Chromatogr B. 2014;972:48–52. doi: 10.1016/j.jchromb.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 65.Williams A, Read EK, Agarabi CD, Lute S, Brorson KA. J Chromatogr B. 2017;1046:122–130. doi: 10.1016/j.jchromb.2017.01.021. [DOI] [PubMed] [Google Scholar]
- 66.Birch JR, Racher AJ. Adv Drug Deliv Rev. 2006;58:671–685. doi: 10.1016/j.addr.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 67.Berkowitz SA, Engen JR, Mazzeo JR, Jones GB. Nat Rev Drug Discov. 2012;11:527–540. doi: 10.1038/nrd3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Q, Luo G, Ou J, Yeung WSB. J Chromatogr A. 1999;848:139–148. doi: 10.1016/s0021-9673(99)00413-6. [DOI] [PubMed] [Google Scholar]
- 69.de Frutos M, Paliwal SK, Regnier FE. Anal Chem. 1993;65:2159–2163. doi: 10.1021/ac00063a040. [DOI] [PubMed] [Google Scholar]
- 70.Shen H, Aspinwall CA, Kennedy RT. J Chromatogr B. 1997;689:295–303. doi: 10.1016/s0378-4347(96)00336-2. [DOI] [PubMed] [Google Scholar]
- 71.Holtzapple CK, Pishko EJ, Stanker LH. Anal Chem. 2000;72:4148–4153. doi: 10.1021/ac000065k. [DOI] [PubMed] [Google Scholar]
- 72.Pfaunmiller EL, Anguizola JA, Milanuk ML, Carter N, Hage DS. J Chromatogr B. 2016;1021:91–100. doi: 10.1016/j.jchromb.2015.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cassidy SA, Janis LJ, Regnier FE. Anal Chem. 1992;64:1973–1977. doi: 10.1021/ac00041a036. [DOI] [PubMed] [Google Scholar]
- 74.Smuc T, Ahn IY, Ulrich H. J Pharm Biomed Anal. 2013;81–82:210–217. doi: 10.1016/j.jpba.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 75.Ma H, Liu J, Ali MM, Mahmood MAI, Labanieh L, Lu M, Iqbal SM, Zhang Q, Zhao W, Wan Y. Chem Soc Rev. 2015;44:1240–1256. doi: 10.1039/c4cs00357h. [DOI] [PubMed] [Google Scholar]
- 76.Proske D, Blank M, Buhmann R, Resch A. Appl Microbiol Biotechnol. 2005;69:367–374. doi: 10.1007/s00253-005-0193-5. [DOI] [PubMed] [Google Scholar]
- 77.Balamurugan S, Obubuafo A, Soper SA, Spivak DA. Anal Bioanal Chem. 2008;390:1009–1021. doi: 10.1007/s00216-007-1587-2. [DOI] [PubMed] [Google Scholar]
- 78.Zhao Q, Wu M, Chris Le X, Li XF. Trends Anal Chem. 2012;41:46–57. [Google Scholar]
- 79.Bouchard PR, Hutabarat RM, Thompson KM. Annu Rev Pharmacol Toxicol. 2010;50:237–257. doi: 10.1146/annurev.pharmtox.010909.105547. [DOI] [PubMed] [Google Scholar]
- 80.Wu YX, Kwon YJ. Methods. 2016;106:21–28. doi: 10.1016/j.ymeth.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 81.Deng Q, German I, Buchanan D, Kennedy RT. Anal Chem. 2001;73:5415–5421. doi: 10.1021/ac0105437. [DOI] [PubMed] [Google Scholar]
- 82.Deng Q, Watson CJ, Kennedy RT. J Chromatogr A. 2003;1005:123–130. doi: 10.1016/s0021-9673(03)00812-4. [DOI] [PubMed] [Google Scholar]
- 83.Scriba GKE. Chromatographia. 2012;75:815–838. [Google Scholar]
- 84.Ruta J, Ravelet C, Desire J, Decout JL, Peyrin E. Anal Bioanal Chem. 2008;390:1051–1057. doi: 10.1007/s00216-007-1552-0. [DOI] [PubMed] [Google Scholar]
- 85.Michaud M, Jourdan E, Villet A, Ravel A, Grosset C, Peyrin E. J Am Chem Soc. 2003;125:8672–8679. doi: 10.1021/ja034483t. [DOI] [PubMed] [Google Scholar]
- 86.Ravelet C, Boulkedid R, Ravel A, Grosset C, Villet A, Fize J, Peyrin E. J Chromatogr A. 2005;1076:62–70. doi: 10.1016/j.chroma.2005.03.132. [DOI] [PubMed] [Google Scholar]
- 87.Romig TS, Bell C, Drolet DW. J Chromatogr B. 1999;731:275–284. [PubMed] [Google Scholar]
- 88.Zhao Q, Li XF, Shao Y, Le XC. Anal Chem. 2008;80:7586–7593. doi: 10.1021/ac801206s. [DOI] [PubMed] [Google Scholar]
- 89.Zhao Q, Li XF, Le XC. Anal Chem. 2008;80:3915–3920. doi: 10.1021/ac702567x. [DOI] [PubMed] [Google Scholar]
- 90.Han B, Zhao C, Yin J, Wang H. J Chromatogr B. 2012;903:112–117. doi: 10.1016/j.jchromb.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 91.Deng N, Liang Z, Liang Y, Sui Z, Zhang L, Wu Q, Yang K, Zhang L, Zhang Y. Anal Chem. 2012;84:10186–10190. doi: 10.1021/ac302779u. [DOI] [PubMed] [Google Scholar]
- 92.Connor AC, McGown LB. J Chromatogr A. 2006;1111:115–119. doi: 10.1016/j.chroma.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 93.Nguyen TH, Pei R, Stojanovic M, Lin Q. Microfluid Nanofluidics. 2009;6:479–487. doi: 10.1007/s10404-012-0993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chung WJ, Kim MS, Cho S, Park SS, Kim JH, Kim YK, Kim BG, Lee YS. Electrophoresis. 2005;26:694–702. doi: 10.1002/elps.200410005. [DOI] [PubMed] [Google Scholar]
- 95.Vellard M. Curr Opin Biotechnol. 2003;14:444–450. doi: 10.1016/s0958-1669(03)00092-2. [DOI] [PubMed] [Google Scholar]
- 96.Moehlenbrock MJ, Minteer SD. In: Enzyme Stabilization and Immobilization. Minteer SD, editor. Springer; New York: 2010. pp. 1–9. [Google Scholar]
- 97.Maximov V, Reukov V, Vertegel AA. J Drug Deliv Sci Technol. 2009;19:311–320. [Google Scholar]
- 98.Bowers-komro DM, Yamada Y, Mccormick DB. Biochemistry. 1989;28:8439–8446. doi: 10.1021/bi00447a025. [DOI] [PubMed] [Google Scholar]
- 99.Kuester M, Becker GL, Lindberg I, Steinmetzer T, Than ME. Biol Chem. 2011;392:973–981. doi: 10.1515/BC.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Homandberg GA, Litwiller RD, Peanasky RJ. Arch Biochem Biophys. 1989;270:153–161. doi: 10.1016/0003-9861(89)90017-9. [DOI] [PubMed] [Google Scholar]
- 101.Martzen MR, McMullen BA, Smith NE, Fujikawa K, Peanasky RJ. Biochemistry. 1990;29:7366–7372. doi: 10.1021/bi00484a003. [DOI] [PubMed] [Google Scholar]
- 102.Zaborsky OR. Methods Enzymol. 1976;44:317–332. doi: 10.1016/s0076-6879(76)44026-0. [DOI] [PubMed] [Google Scholar]
- 103.Friedberg F, Rhoads AR. In: Handbook of Affinity Chromatography. 2. Hage DS, editor. CRC Press; Boca Raton: 2005. pp. 313–346. [Google Scholar]
- 104.Hage DS, Bian M, Burks R, Karle E, Ohnmacht C, Wa C. In: Handbook of Affinity Chromatography. 2. Hage DS, editor. CRC Press; Boca Raton: 2005. pp. 101–126. [Google Scholar]
- 105.John Lough W, Wainer IW, Patel S. In: Handbook of Affinity Chromatography. 2. Hage DS, editor. CRC Press; Boca Raton: 2005. pp. 571–592. [Google Scholar]
- 106.Haginaka J. J Chromatogr B. 2008;875:12–19. doi: 10.1016/j.jchromb.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 107.Calleri E, Massolini G, Loiodice F, Fracchiolla G, Temporini C, Félix G, Tortorella P, Caccialanza G. J Chromatogr A. 2002;958:131–140. doi: 10.1016/s0021-9673(02)00403-x. [DOI] [PubMed] [Google Scholar]
- 108.Calleri E, Massolini G, Lubda D, Temporini C, Loiodice F, Caccialanza G. J Chromatogr A. 2004;1031:93–100. doi: 10.1016/j.chroma.2003.08.076. [DOI] [PubMed] [Google Scholar]
- 109.Massolini G, Calleri E, Lavecchia A, Loiodice F, Lubda D, Temporini C, Fracchiolla G, Tortorella P, Novellino E, Caccialanza G. Anal Chem. 2003;75:535–542. doi: 10.1021/ac0204193. [DOI] [PubMed] [Google Scholar]
- 110.Stoffer B, Frandsen TP, Busk PK, Schneider P, Svendsen I, Svensson B. Biochem J. 1993;292:197–202. doi: 10.1042/bj2920197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Strandberg A, Nyström A, Behr S, Karlsson A. Chromatographia. 1999;50:215–222. [Google Scholar]
- 112.Zheng Y, Wang X, Ji Y. Talanta. 2012;91:7–17. doi: 10.1016/j.talanta.2012.01.039. [DOI] [PubMed] [Google Scholar]
- 113.Fanali S, Caponecchi G, Aturki Z. J Microcolumn Sep. 1997;9:9–14. [Google Scholar]
- 114.Ståhlberg J, Henriksson H, Divne C, Isaksson R, Pettersson G, Johansson G, Jones TA. J Mol Biol. 2001;305:79–93. doi: 10.1006/jmbi.2000.4237. [DOI] [PubMed] [Google Scholar]
- 115.Bartolini M, Cavrini V, Andrisano V. J Chromatogr A. 2007;1144:102–110. doi: 10.1016/j.chroma.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 116.Forsberg EM, Green A, JR, Brennan JD. Anal Chem. 2011;83:5230–5236. doi: 10.1021/ac200534t. [DOI] [PubMed] [Google Scholar]
- 117.Forsberg EM, Sicard C, Brennan JD. Annu Rev Anal Chem. 2014;7:337–359. doi: 10.1146/annurev-anchem-071213-020241. [DOI] [PubMed] [Google Scholar]
- 118.Kim HS, Wainer IW. J Chromatogr B. 2005;823:158–166. doi: 10.1016/j.jchromb.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 119.Hodgson RJ, Besanger TR, Brook MA, Brennan JD. Anal Chem. 2005;77:7512–7519. doi: 10.1021/ac050761q. [DOI] [PubMed] [Google Scholar]
- 120.Luckarift HR. J Liq Chromatogr Relat Technol. 2008;31:1568–1592. [Google Scholar]
- 121.Matosevic S, Szita N, Baganz F. J Chem Technol Biotechnol. 2011;86:325–334. [Google Scholar]
- 122.Girelli AM, Mattei E. J Chromatogr B. 2005;819:3–16. doi: 10.1016/j.jchromb.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 123.Hong T, Chen X, Xu Y, Cui X, Bai R, Jin C. J Chromatogr A. 2016;1456:249–256. doi: 10.1016/j.chroma.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 124.Yuan H, Zhang L, Zhang Y. J Chromatogr A. 2014;1371:48–57. doi: 10.1016/j.chroma.2014.10.067. [DOI] [PubMed] [Google Scholar]
- 125.Liang W, Hou Z, Wang H, Xu W, Wang W. Chromatographia. 2015;78:763–773. [Google Scholar]
- 126.Wang X, Li K, Adams E, Van Schepdael A. Electrophoresis. 2014;35:119–127. doi: 10.1002/elps.201300294. [DOI] [PubMed] [Google Scholar]
- 127.Scriba GKE, Belal F. Chromatographia. 2015;78:947–970. [Google Scholar]
- 128.Peumans WJ, Van Damme EJ. Plant Physiol. 1995;109:347–352. doi: 10.1104/pp.109.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Alley WR, Mann BF, Novotny MV. Chem Rev. 2013;113:2668–2732. doi: 10.1021/cr3003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Van Damme EJM. Handbook of Plant Lectins : Properties and Biomedical Applications. John Wiley; Chichester: 1998. [Google Scholar]
- 131.Kobayashi Y, Tateno H, Ogawa H, Yamamoto K, Hirabayashi J. In: Lectins. Hirabayashi J, editor. Springer; New York: 2014. pp. 555–577. [DOI] [PubMed] [Google Scholar]
- 132.Bergström M, Nilsson M, Isaksson R, Rydén I, Påhlsson P, Ohlson S. J Chromatogr B. 2004;809:323–329. doi: 10.1016/j.jchromb.2004.06.042. [DOI] [PubMed] [Google Scholar]
- 133.Bedair M, El Rassi Z. J Chromatogr A. 2004;1044:177–186. doi: 10.1016/j.chroma.2004.03.080. [DOI] [PubMed] [Google Scholar]
- 134.Luo R, Archer-Hartmann SA, Holland LA. Anal Chem. 2010;82:1228–1233. doi: 10.1021/ac902052m. [DOI] [PubMed] [Google Scholar]
- 135.Okanda FM, El Rassi Z. Electrophoresis. 2006;27:1020–1030. doi: 10.1002/elps.200500766. [DOI] [PubMed] [Google Scholar]
- 136.Fukushima E, Yagi Y, Yamamoto S, Nakatani Y, Kakehi K, Hayakawa T, Suzuki S. J Chromatogr A. 2012;1246:84–89. doi: 10.1016/j.chroma.2012.02.052. [DOI] [PubMed] [Google Scholar]
- 137.Madera M, Mechref Y, Novotny MV. Anal Chem. 2005;77:4081–4090. doi: 10.1021/ac050222l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang Z, Hancock WS. J Chromatogr A. 2004;1053:79–88. [PubMed] [Google Scholar]
- 139.Durham M, Regnier FE. J Chromatogr A. 2006;1132:165–173. doi: 10.1016/j.chroma.2006.07.070. [DOI] [PubMed] [Google Scholar]
- 140.Yang Z, Hancock WS. J Chromatogr A. 2005;1070:57–64. doi: 10.1016/j.chroma.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 141.Ahn J-M, Kim B-G, Yu M-H, Lee I-K, Cho J-Y. PROTEOMICS – Clin Appl. 2010;4:644–653. doi: 10.1002/prca.200900196. [DOI] [PubMed] [Google Scholar]
- 142.Lee LY, Hincapie M, Packer N, Baker MS, Hancock WS, Fanayan S. J Sep Sci. 2012;35:2445–2452. doi: 10.1002/jssc.201200049. [DOI] [PubMed] [Google Scholar]
- 143.Dai L, Liu Y, He J, Flack CG, Talsma CE, Crowley JG, Muraszko KM, Fan X, Lubman DM. Proteomics. 2011;11:4021–4028. doi: 10.1002/pmic.201100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kullolli M, Hancock WS, Hincapie M. Anal Chem. 2010;82:115–120. doi: 10.1021/ac9013308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gbormittah FO, Hincapie M, Hancock WS. Bioanalysis. 2014;6:2537–2548. doi: 10.4155/bio.14.217. [DOI] [PubMed] [Google Scholar]
- 146.Gbormittah FO, Lee LY, Taylor K, Hancock WS, Iliopoulos O. J Proteome Res. 2014;13:4889–4900. doi: 10.1021/pr500591e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zeng Z, Hincapie M, Pitteri SJ, Hanash S, Schalkwijk J, Hogan JM, Wang H, Hancock WS. Anal Chem. 2011;83:4845–4854. doi: 10.1021/ac2002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jung K, Cho W. Anal Chem. 2013;85:7125–7132. doi: 10.1021/ac400653z. [DOI] [PubMed] [Google Scholar]
- 149.Qiu R, Regnier FE. Anal Chem. 2005;77:2802–2809. doi: 10.1021/ac048751x. [DOI] [PubMed] [Google Scholar]
- 150.Qiu R, Regnier FE. Anal Chem. 2005;77:7225–7231. doi: 10.1021/ac050554q. [DOI] [PubMed] [Google Scholar]
- 151.Kullolli M, Hancock WS, Hincapie M. J Sep Sci. 2008;31:2733–2739. doi: 10.1002/jssc.200800233. [DOI] [PubMed] [Google Scholar]
- 152.Zeng Z, Hincapie M, Haab BB, Hanash S, Pitteri SJ, Kluck S, Hogan JM, Kennedy J, Hancock WS. J Chromatogr A. 2010;1217:3307–3315. doi: 10.1016/j.chroma.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Madera M, Mechref Y, Klouckova I, Novotny MV. J Chromatogr B. 2007;845:121–137. doi: 10.1016/j.jchromb.2006.07.067. [DOI] [PubMed] [Google Scholar]
- 154.Madera M, Mechref Y, Klouckova I, Novotny MV. J Proteome Res. 2006;5:2348–2363. doi: 10.1021/pr060169x. [DOI] [PubMed] [Google Scholar]
- 155.Mann BF, Mann AKP, Skrabalak SE, Novotny MV. Anal Chem. 2013;85:1905–1912. doi: 10.1021/ac303274w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bedair M, El Rassi Z. J Chromatogr A. 2005;1079:236–245. doi: 10.1016/j.chroma.2005.02.084. [DOI] [PubMed] [Google Scholar]
- 157.Feng S, Yang N, Pennathur S, Goodison S, Lubman DM. Anal Chem. 2009;81:3776–3783. doi: 10.1021/ac900085k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Alwael H, Connolly D, Clarke P, Thompson R, Twamley B, O’Connor B, Paull B. Analyst. 2011;136:2619–2628. doi: 10.1039/c1an15137a. [DOI] [PubMed] [Google Scholar]
- 159.Alla AJ, d’ Andrea FB, Bhattarai JK, Cooper JA, Tan YH, Demchenko AV, Stine KJ. J Chromatogr A. 2015;1423:19–30. doi: 10.1016/j.chroma.2015.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Peters T. All About Albumin: Biochemistry Genetics and Medical Applications. 1. Academic Press; San Diego: 1996. [Google Scholar]
- 161.Spector AA, John K, Fletcher JE. J Lipid Res. 1969;10:56–67. [PubMed] [Google Scholar]
- 162.Arroyo V, García-Martinez R, Salvatella X. J Hepatol. 2014;61:396–407. doi: 10.1016/j.jhep.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 163.Haginaka J. J Chromatogr A. 2001;906:253–273. doi: 10.1016/s0021-9673(00)00504-5. [DOI] [PubMed] [Google Scholar]
- 164.Fournier T, Medjoubi N, Porquet D. Biochim Biophys Acta. 2000;1482:157–171. doi: 10.1016/s0167-4838(00)00153-9. [DOI] [PubMed] [Google Scholar]
- 165.Kremer JM, Wilting J, Janssen LH. Pharmacol Rev. 1988;40:1–47. [PubMed] [Google Scholar]
- 166.Patel S, Wainer IW, Lough WJ. In: Handbook of Affinity Chromatography. 2. Hage DS, editor. CRC Press; Boca Raton: 2005. pp. 663–684. [Google Scholar]
- 167.Harmony JAK, Aleson AL, McCarthy BM. In: Biochemistry and Biology of Plasma Lipoproteins. Scanu AM, Spector AA, editors. Marcel Dekker Inc; New York: 1986. pp. 403–430. [Google Scholar]
- 168.Mbewu AD, Durrington PN. Atherosclerosis. 1990;85:1–14. doi: 10.1016/0021-9150(90)90177-k. [DOI] [PubMed] [Google Scholar]
- 169.Durrington PN. Lipoproteins and lipids. Wright; London: 1989. [Google Scholar]
- 170.Havel RJ, Kane JP. In: The Metabolic and Molecular Basis of Inherited Disease. 7. Scriver CR, Beaudet AL, Sly WS, Valle D, editors. McGraw-Hill; New York: 1995. pp. 1129–1138. [Google Scholar]
- 171.Skipski VR. In: Blood Lipids and Lipoproteins: Quantitation, Composition, and Metabolism. Nelson GJ, editor. John Wiley; New York: 1972. pp. 471–583. [Google Scholar]
- 172.Barklay M. In: Blood Lipids and Lipoproteins: Quantitation, Composition, and Metabolism. Nelson GJ, editor. John Wiley; New York: 1972. pp. 587–603. [Google Scholar]
- 173.Jonas A. In: Biochemistry of Lipids, Lipoproteins and Membranes. Vance JE, Vance D, editors. Elsevier; Amsterdam: 2002. pp. 483–504. [Google Scholar]
- 174.Fruchart JC. Human plasma lipoproteins. Walter De Gruyter Inc; Berlin: 1989. [Google Scholar]
- 175.Wasan KM, Cassidy SM. J Pharm Sci. 1998;87:411–424. doi: 10.1021/js970407a. [DOI] [PubMed] [Google Scholar]
- 176.Domenici E, Bertucci C, Salvadori P, Felix G, Cahagne I, Motellier S, Wainer IW. Chromatographia. 1990;29:170–176. [Google Scholar]
- 177.Kaliszan R, Noctor TA, Wainer IW. Mol Pharmacol. 1992;42:512–517. [PubMed] [Google Scholar]
- 178.Noctor TA, Pham CD, Kaliszan R, Wainer IW. Mol Pharmacol. 1992;42:506–511. [PubMed] [Google Scholar]
- 179.Allenmark S, Andersson S. Chirality. 1989;1:154–160. [Google Scholar]
- 180.Loun B, Hage DS. J Chromatogr B. 1992;579:225–235. [PubMed] [Google Scholar]
- 181.Yang J, Hage DS. J Chromatogr A. 1993;645:241–250. doi: 10.1016/0021-9673(93)83383-4. [DOI] [PubMed] [Google Scholar]
- 182.Chu Y-Q, Wainer IW. Pharm Res. 1988;5:680–683. doi: 10.1023/a:1015991324316. [DOI] [PubMed] [Google Scholar]
- 183.Pfaunmiller EL, Hartmann M, Dupper CM, Soman S, Hage DS. J Chromatogr A. 2012;1269:198–207. doi: 10.1016/j.chroma.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mallik R, Hage DS. J Pharm Biomed Anal. 2008;46:820–830. doi: 10.1016/j.jpba.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lu J, Ye F, Zhang A, Wei Z, Peng Y, Zhao S. J Sep Sci. 2011;34:2329–2336. doi: 10.1002/jssc.201100102. [DOI] [PubMed] [Google Scholar]
- 186.Hong T, Zheng Y, Hu W, Ji Y. Anal Biochem. 2014;464:43–50. doi: 10.1016/j.ab.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 187.Breton D, Buret D, Clair P, Lafosse M. J Chromatogr A. 2005;1088:104–109. doi: 10.1016/j.chroma.2005.02.073. [DOI] [PubMed] [Google Scholar]
- 188.Enquist M, Hermansson J. Chirality. 2004;1:209–215. doi: 10.1002/chir.530010306. [DOI] [PubMed] [Google Scholar]
- 189.Locatelli I, Kmetec V, Mrhar A, Grabnar I. J Chromatogr B. 2005;818:191–198. doi: 10.1016/j.jchromb.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 190.Sandström R, Lennernäs H, Öhlén K, Karlsson A. J Pharm Biomed Anal. 1999;21:43–49. doi: 10.1016/s0731-7085(99)00093-x. [DOI] [PubMed] [Google Scholar]
- 191.Soares R, Singh AK, Kedor-Hackmann ERM, Santoro MIRM. J AOAC Int. 2009;92:1663–1672. [PubMed] [Google Scholar]
- 192.Etter ML, George S, Graybiel K, Eichhorst J, Lehotay DC. Clin Biochem. 2005;38:1095–1102. doi: 10.1016/j.clinbiochem.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 193.Du J, Ma Z, Zhang Y, Wang T, Chen X, Zhong D. J Pharm Biomed Anal. 2013;86:182–188. doi: 10.1016/j.jpba.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 194.Kawabata K, Samata N, Urasaki Y, Fukazawa I, Uchida N, Uchida E, Yasuhara H. J Chromatogr B. 2007;852:389–397. doi: 10.1016/j.jchromb.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 195.Liu K, Dai X, Zhong D, Chen X. J Chromatogr B. 2008;864:129–136. doi: 10.1016/j.jchromb.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 196.Liu K, Zhong D, Chen X. J Chromatogr B. 2010;878:2415–2420. doi: 10.1016/j.jchromb.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 197.Xuan H, Hage DS. J Sep Sci. 2006;29:1412–1422. doi: 10.1002/jssc.200600051. [DOI] [PubMed] [Google Scholar]
- 198.Mallik R, Xuan H, Hage DS. J Chromatogr A. 2007;1149:294–304. doi: 10.1016/j.chroma.2007.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Karlsson A, Aspegren A. Chromatographia. 1998;47:189–196. [Google Scholar]
- 200.Loeser E, Yowell G, Drumm P. J Liq Chromatogr Rel Technol. 2006;29:2625–2640. [Google Scholar]
- 201.Matsuda R, Li Z, Zheng X, Hage DS. J Chromatogr A. 2015;1408:133–144. doi: 10.1016/j.chroma.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Joseph KS, Hage DS. J Chromatogr B. 2010;878:1590–1598. doi: 10.1016/j.jchromb.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Xuan H, Joseph KS, Wa C, Hage DS. J Sep Sci. 2010;33:2294–2301. doi: 10.1002/jssc.201000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hage DS. J Chromatogr B. 2002;768:3–30. doi: 10.1016/s0378-4347(01)00482-0. [DOI] [PubMed] [Google Scholar]
- 205.Hage DS, Jackson A, Sobansky MR, Schiel JE, Yoo MJ, Joseph KS. J Sep Sci. 2009;32:835–53. doi: 10.1002/jssc.200800640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Loun B, Hage DS. Anal Chem. 1994;66:3814–3822. doi: 10.1021/ac00093a043. [DOI] [PubMed] [Google Scholar]
- 207.Yang J, Hage DS. J Chromatogr A. 1996;725:273–285. doi: 10.1016/0021-9673(95)01009-2. [DOI] [PubMed] [Google Scholar]
- 208.Tweed SA, Loun B, Hage DS. Anal Chem. 1997;69:4790–4798. doi: 10.1021/ac970565m. [DOI] [PubMed] [Google Scholar]
- 209.Kim HS, Hage DS. J Chromatogr B. 2005;816:57–66. doi: 10.1016/j.jchromb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 210.Yoo MJ, Smith QR, Hage DS. J Chromatogr B. 2009;877:1149–1154. doi: 10.1016/j.jchromb.2009.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Soman S, Yoo MJ, Jang YJ, Hage DS. J Chromatogr B. 2010;878:705–708. doi: 10.1016/j.jchromb.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Chen J, Ohnmacht C, Hage DS. J Chromatogr B. 2004;809:137–145. doi: 10.1016/j.jchromb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 213.Mallik R, Yoo MJ, Chen S, Hage DS. J Chromatogr B. 2008;876:69–75. doi: 10.1016/j.jchromb.2008.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Joseph KS, Moser AC, Basiaga SBG, Schiel JE, Hage DS. J Chromatogr A. 2009;1216:3492–3500. doi: 10.1016/j.chroma.2008.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Conrad ML, Moser AC, Hage DS. J Sep Sci. 2009;32:1145–1155. doi: 10.1002/jssc.200800567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sobansky MR, Hage DS. Adv Med Biol. 2012;53:199–216. [PMC free article] [PubMed] [Google Scholar]
- 217.Sobansky MR, Hage DS. Anal Bioanal Chem. 2014;406:6203–6211. doi: 10.1007/s00216-014-8081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Zheng X, Li Z, Beeram S, Podariu M, Matsuda R, Pfaunmiller EL, White CJ, Carter N, Hage DS. J Chromatogr B. 2014;968:49–63. doi: 10.1016/j.jchromb.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Beeram S, Bi C, Zheng X, Hage DS. J Chromatogr A. 2017;1497:92–101. doi: 10.1016/j.chroma.2017.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Mallik R, Yoo MJ, Briscoe CJ, Hage DS. J Chromatogr A. 2010;1217:2796–2803. doi: 10.1016/j.chroma.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zheng X, Li Z, Podariu MI, Hage DS. Anal Chem. 2014;86:6454–6460. doi: 10.1021/ac501031y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Bi C, Zheng X, Hage DS. J Chromatogr A. 2016;1432:49–57. doi: 10.1016/j.chroma.2015.12.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zheng X, Yoo MJ, Hage DS. Analyst. 2013;138:6262–5. doi: 10.1039/c3an01315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Clarke W, Chowdhuri R, Hage DS. Anal Chem. 2001;73:2157–2164. doi: 10.1021/ac0009752. [DOI] [PubMed] [Google Scholar]
- 225.Zheng X, Matsuda R, Hage DS. J Chromatogr A. 2014;1371:82–89. doi: 10.1016/j.chroma.2014.10.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zheng X, Bi C, Brooks M, Hage DS. Anal Chem. 2015;87:11187–11194. doi: 10.1021/acs.analchem.5b03007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zheng X, Podariu M, Matsuda R, Hage DS. Anal Bioanal Chem. 2016;408:131–140. doi: 10.1007/s00216-015-9082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Stewart BH, Helen Chan O. J Pharm Sci. 1998;87:1471–1478. doi: 10.1021/js980262n. [DOI] [PubMed] [Google Scholar]
- 229.Ong S, Liu H, Pidgeon C. J Chromatogr A. 1996;728:113–128. doi: 10.1016/0021-9673(95)00837-3. [DOI] [PubMed] [Google Scholar]
- 230.Tsopelas F, Vallianatou T, Tsantili-Kakoulidou A. Expert Opin Drug Discov. 2016;11:473–488. doi: 10.1517/17460441.2016.1160886. [DOI] [PubMed] [Google Scholar]
- 231.Yang CY, Cai SJ, Liu H, Pidgeon C. Adv Drug Deliv Rev. 1997;23:229–256. [Google Scholar]
- 232.Ong S, Liu H, Qiu X, Bhat G, Pidgeon C. Anal Chem. 1995;67:755–762. doi: 10.1021/ac00100a011. [DOI] [PubMed] [Google Scholar]
- 233.Pidgeon C, Venkataram UV. Anal Biochem. 1989;176:36–47. doi: 10.1016/0003-2697(89)90269-8. [DOI] [PubMed] [Google Scholar]
- 234.De Vrieze M, Verzele D, Szucs R, Sandra P, Lynen F. Anal Bioanal Chem. 2014;406:6179–6188. doi: 10.1007/s00216-014-8054-7. [DOI] [PubMed] [Google Scholar]
- 235.Moaddel R, Lu L, Baynham M, Wainer IW. J Chromatogr B. 2002;768:41–53. doi: 10.1016/s0378-4347(01)00484-4. [DOI] [PubMed] [Google Scholar]
- 236.Zhang Y, Xiao Y, Kellar KJ, Wainer IW. Anal Biochem. 1998;264:22–25. doi: 10.1006/abio.1998.2828. [DOI] [PubMed] [Google Scholar]
- 237.Wainer IW, Zhang Y, Xiao Y, Kellar KJ. J Chromatogr B. 1999;724:65–72. doi: 10.1016/s0378-4347(98)00579-9. [DOI] [PubMed] [Google Scholar]
- 238.Zhang Y, Leonessa F, Clarke R, Wainer IW. J Chromatogr B. 2000;739:33–37. doi: 10.1016/s0378-4347(99)00384-9. [DOI] [PubMed] [Google Scholar]
- 239.Lu L, Leonessa F, Clarke R, Wainer IW. Mol Pharmacol. 2001;59:62–68. doi: 10.1124/mol.59.1.62. [DOI] [PubMed] [Google Scholar]
- 240.Beigi F, Qing Y, Lundahl P. J Chromatogr A. 1995;704:315–321. doi: 10.1016/0021-9673(95)00214-8. [DOI] [PubMed] [Google Scholar]
- 241.Lundahl P, Beigi F. Adv Drug Deliv Rev. 1997;23:221–227. [Google Scholar]
- 242.Liu X-Y, Nakamura C, Yang Q, Kamo N, Miyake J. J Chromatogr A. 2002;961:113–118. doi: 10.1016/s0021-9673(02)00505-8. [DOI] [PubMed] [Google Scholar]
- 243.Mao X, Kong L, Luo Q, Li X, Zou H. J Chromatogr B. 2002;779:331–339. doi: 10.1016/s1570-0232(02)00403-8. [DOI] [PubMed] [Google Scholar]
- 244.Zhang C, Li J, Xu L, Shi ZG. J Chromatogr A. 2012;1233:78–84. doi: 10.1016/j.chroma.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 245.Wang Y, Kong L, Lei X, Hu L, Zou H, Welbeck E, Bligh SWA, Wang Z. J Chromatogr A. 2009;1216:2185–2191. doi: 10.1016/j.chroma.2008.05.074. [DOI] [PubMed] [Google Scholar]
- 246.Moravcová D, Planeta J, Wiedmer SK. J Chromatogr A. 2013;1317:159–166. doi: 10.1016/j.chroma.2013.08.031. [DOI] [PubMed] [Google Scholar]
- 247.Chen X, Xia Y, Lu Y, Liang J. J Pharm Biomed Anal. 2011;54:406–410. doi: 10.1016/j.jpba.2010.08.028. [DOI] [PubMed] [Google Scholar]
- 248.Chen X, Deng Y, Xue Y, Liang J. J Pharm Biomed Anal. 2012;70:194–201. doi: 10.1016/j.jpba.2012.06.030. [DOI] [PubMed] [Google Scholar]
- 249.Sheng L-H, Li S-L, Kong L, Chen X-G, Mao X-Q, Su X-Y, Zou H-F, Li P. J Pharm Biomed Anal. 2005;38:216–224. doi: 10.1016/j.jpba.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 250.Szumski M, Grzywiński D, Buszewski B. J Chromatogr A. 2014;1373:114–123. doi: 10.1016/j.chroma.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 251.Al-Haj MA, Haber P, Kaliszan R, Buszewski B, Jezierska M, Chilmonzyk Z. J Pharm Biomed Anal. 1998;18:721–728. doi: 10.1016/s0731-7085(98)00287-8. [DOI] [PubMed] [Google Scholar]
- 252.Grzywiński D, Szumski M, Buszewski B. J Chromatogr A. 2015;1408:145–150. doi: 10.1016/j.chroma.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 253.Moravcová D, Carrasco-Correa EJ, Planeta J, Lämmerhofer M, Wiedmer SK. J Chromatogr A. 2015;1402:27–35. doi: 10.1016/j.chroma.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 254.Wang Q, Peng K, Chen W, Cao Z, Zhu P, Zhao Y, Wang Y, Zhou H, Jiang Z. J Chromatogr A. 2017;1479:97–106. doi: 10.1016/j.chroma.2016.11.046. [DOI] [PubMed] [Google Scholar]
- 255.Armstrong DW, Ward TJ, Armstrong RD, Beesley TE. Science. 1986;232:1132–1135. doi: 10.1126/science.3704640. [DOI] [PubMed] [Google Scholar]
- 256.Ward TJ, Ward KD. Anal Chem. 2012;84:626–635. doi: 10.1021/ac202892w. [DOI] [PubMed] [Google Scholar]
- 257.Shen J, Ikai T, Okamoto Y. J Chromatogr A. 2014;1363:51–61. doi: 10.1016/j.chroma.2014.06.042. [DOI] [PubMed] [Google Scholar]
- 258.Chankvetadze B. J Chromatogr A. 2012;1269:26–51. doi: 10.1016/j.chroma.2012.10.033. [DOI] [PubMed] [Google Scholar]
- 259.Szejtli J. Chem Rev. 1998;98:1743–1754. doi: 10.1021/cr970022c. [DOI] [PubMed] [Google Scholar]
- 260.Crini G. Chem Rev. 2014;114:10940–10975. doi: 10.1021/cr500081p. [DOI] [PubMed] [Google Scholar]
- 261.Hinze WL, Riehl TE, Armstrong DW, DeMond W, Alak A, Ward T. Anal Chem. 1985;57:237–242. [Google Scholar]
- 262.Risley DS, Strege MA. Anal Chem. 2000;72:1736–1739. doi: 10.1021/ac9911490. [DOI] [PubMed] [Google Scholar]
- 263.Liu Y, Xue X, Guo Z, Xu Q, Zhang F, Liang X. J Chromatogr A. 2008;1208:133–140. doi: 10.1016/j.chroma.2008.08.079. [DOI] [PubMed] [Google Scholar]
- 264.Wang H, Xu D, Jiang P, Zhang M, Dong X. Analyst. 2010;135:1785–1792. doi: 10.1039/c0an00050g. [DOI] [PubMed] [Google Scholar]
- 265.Zhou J, Tang J, Tang W. Trends Anal Chem. 2015;65:22–29. [Google Scholar]
- 266.Yao X, Tan TTY, Wang Y. J Chromatogr A. 2014;1326:80–88. doi: 10.1016/j.chroma.2013.12.054. [DOI] [PubMed] [Google Scholar]
- 267.Wang R-Q, Ong T-T, Tang W, Ng S-C. Anal Chim Acta. 2012;718:121–129. doi: 10.1016/j.aca.2011.12.063. [DOI] [PubMed] [Google Scholar]
- 268.Lin Y, Zhou J, Tang J, Tang W. J Chromatogr A. 2015;1406:342–346. doi: 10.1016/j.chroma.2015.06.051. [DOI] [PubMed] [Google Scholar]
- 269.Zhao J, Lu X, Wang Y, Lv J. J Chromatogr A. 2015;1381:253–259. doi: 10.1016/j.chroma.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 270.Wang Y, Chen H, Xiao Y, Ng CH, Oh TS, Tan TTY, Ng SC. Nat Protoc. 2011;6:935–942. doi: 10.1038/nprot.2011.340. [DOI] [PubMed] [Google Scholar]
- 271.Kolb HC, Finn MG, Sharpless KB. Angew Chemie Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 272.Guo Z, Lei A, Zhang Y, Xu Q, Xue X, Zhang F, Liang X. Chem Commun. 2007:2491–2493. doi: 10.1039/b701831b. [DOI] [PubMed] [Google Scholar]
- 273.Jin G, Yu D, Guo Z, Yang D, Zhang H, Shen A, Yan J, Liang X. RSC Adv. 2016;6:8584–8587. [Google Scholar]
- 274.Lubda D, Cabrera K, Nakanishi K, Lindner W. Anal Bioanal Chem. 2003;377:892–901. doi: 10.1007/s00216-003-2181-x. [DOI] [PubMed] [Google Scholar]
- 275.Ai F, Wang Y, Chen H, Yang Y, Yang Tan TT, Ng S-C. Analyst. 2013;138:2289–2294. doi: 10.1039/c3an36125j. [DOI] [PubMed] [Google Scholar]
- 276.Barhate CL, Breitbach ZS, Pinto EC, Regalado EL, Welch CJ, Armstrong DW. J Chromatogr A. 2015;1426:241–247. doi: 10.1016/j.chroma.2015.11.056. [DOI] [PubMed] [Google Scholar]
- 277.Zhang Z, Wu M, Wu R, Dong J, Ou J, Zou H. Anal Chem. 2011;83:3616–3622. doi: 10.1021/ac200414r. [DOI] [PubMed] [Google Scholar]
- 278.Zhang Q, Guo J, Wang F, Crommen J, Jiang Z. J Chromatogr A. 2014;1325:147–154. doi: 10.1016/j.chroma.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 279.Sun P, Wang C, Breitbach ZS, Zhang Y, Armstrong DW. Anal Chem. 2009;81:10215–10226. doi: 10.1021/ac902257a. [DOI] [PubMed] [Google Scholar]
- 280.Sun P, Armstrong DW. J Chromatogr A. 2010;1217:4904–4918. doi: 10.1016/j.chroma.2010.04.079. [DOI] [PubMed] [Google Scholar]
- 281.Sun P, Wang C, Padivitage NLT, Nanayakkara YS, Perera S, Qiu H, Zhang Y, Armstrong DW. Analyst. 2011;136:787–800. doi: 10.1039/c0an00653j. [DOI] [PubMed] [Google Scholar]