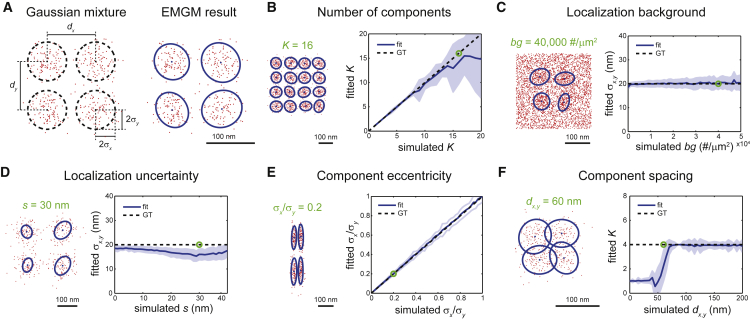
Figure 2.
Evaluation of EMGM using simulated data. (A) On the left is an example of a simulated Gaussian mixture consisting of K = 4 components, each containing 100 localizations, described by a symmetric 2D Gaussian distribution with a SD σx = σy = 20 nm. The Gaussian centers are placed in a square grid with spacing dx,y = 100 nm. On the right is the EMGM result. The red dots symbolize the localizations. The blue dots symbolize the center positions and the blue ellipses symbolize the 2σ error ellipses of the components. (B) On the right, the average number of mixture components correctly identified by EMGM as a function of the simulated K. On the left is an example EMGM result for K = 16. (C) On the right is the average SD σx,y of the mixture components calculated by EMGM as a function of the simulated localization background density bg. On the left is an example EMGM result for bg = 40,000 #/μm2. (D) On the right is the average σx,y calculated by EMGM as a function of the simulated localization uncertainty s. On the left is an example EMGM result for s = 30 nm. (E) On the right is the average eccentricity σx/σy of the mixture components calculated by EMGM as a function of the simulated σx/σy. On the left is an example EMGM result for σx/σy = 0.2. (F) On the right is the average number of mixture components correctly identified by EMGM as a function of the simulated spacing dx,y. On the left is an example EMGM result for dx,y = 60 nm. The simulated Gaussian mixtures in (C–F) consist of K = 4 components, similar to (A). The dashed lines in (B–F) represent the ground truth, and the shaded areas represent the SD (n = 100).