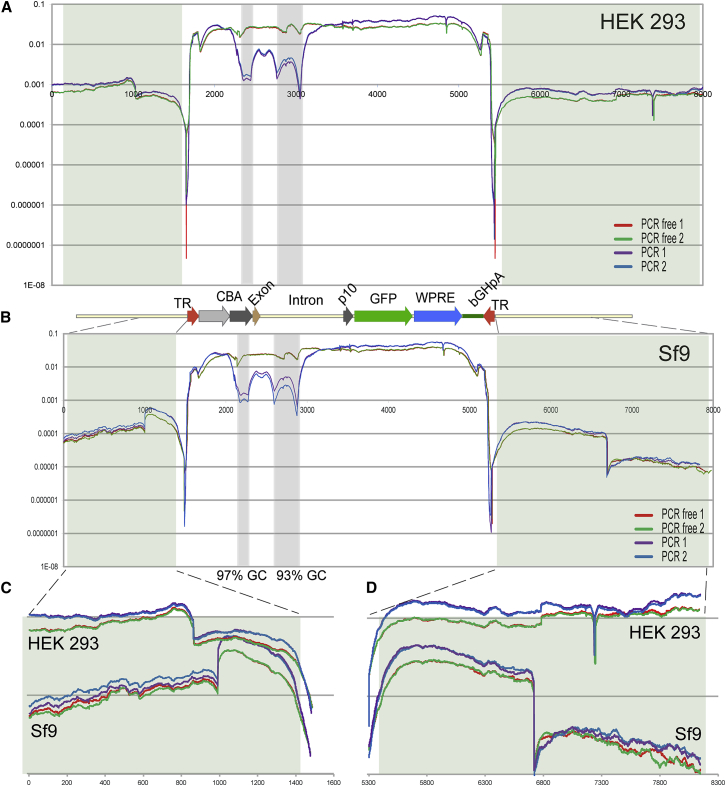
Figure 5.
Distribution of the NGS Reads of the rAAV-Bac-UF26 Cassette and Its Immediate Junctions
(A) Illumina reads coverage of rAAV5-Bac-UF26 cassette packaged in HEK293 cells numerically normalized by dividing the number of reads at each nucleotide position by the total number of reads of the referenced sequence. Sequence coverage graphs for duplicate samples are drawn in different colors: PCR-free, red and green; PCR-based, purple and blue. The drop of coverage between rAAV cassette and the adjacent sequences reflects a graphic representation of the bioinformatics analysis limitations rather than actual reduction in sequence coverage. Shown below the graph is an annotated map of referenced rAAV-Bac-UF26 cassette drawn to the scale of the sequences in (A) and (B). Sequences immediately adjacent to the rAAV cassette in the bacterial plasmid DNA are shaded (green tint). (B) Illumina reads coverage of rAAV5-Bac-UF26 cassette packaged in Sf9 B8 cells. GC-enriched sequences within rAAV cassette are shaded (blue tint). (C) Zoom from (A) and (B) of the sequences immediately adjacent to the left rAAV ITR in both HEK293 and Sf9 B8 cells. (D) Zoom from (A) and (B) of the sequences immediately adjacent to the right rAAV ITR in both HEK293 and Sf9 B8 cells.