Figure 2.
Hyperglycemia-Induced Promoter Hypomethylation Elevates Endothelial MicroRNA-200b
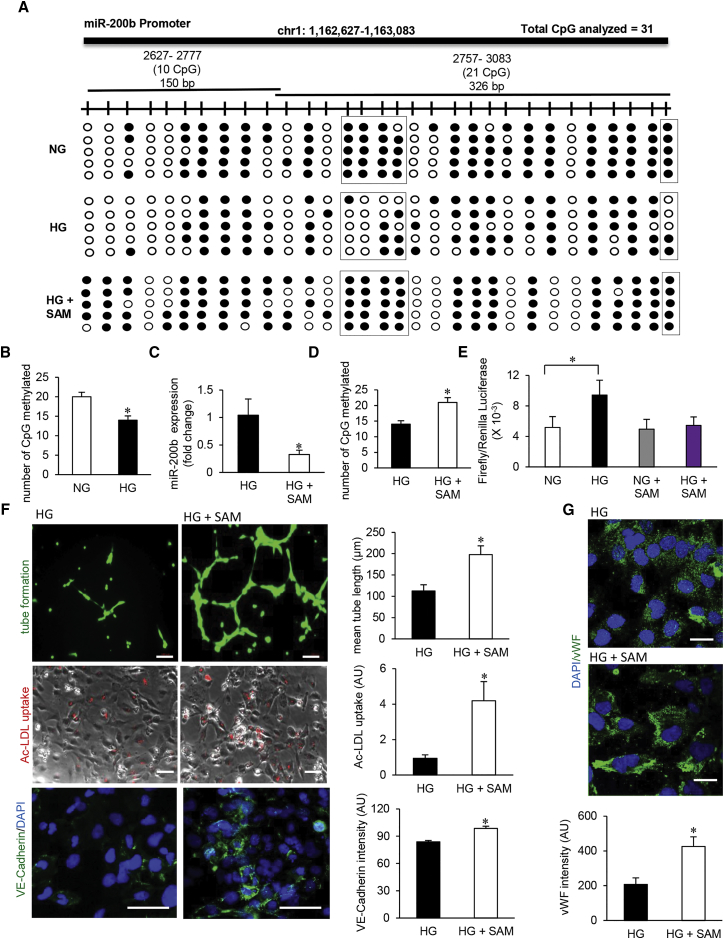
(A) Schematic diagram showing the regions (1, 2) of miR-200b promoter analyzed through bisulfite genomic sequencing of DNA. Methylation profile of the miR-200b promoter in HMECs cultured in normal glucose (NG), exposed to high glucose (HG) and HG supplemented with S-adenosylmethionine (SAM) (80 μM, 48 hr) conditions (methylated CpG, black; unmethylated CpG, white). Number of clones = 5. (B) Total number of methylated CpG sites obtained from bisulfite sequencing analysis. n = 5, *p < 0.05 (Student’s t test). (C) qRT-PCR analysis of miR-200b expression in HMECs treated with SAM under HG condition (80 μM, 48 hr). n = 3, *p < 0.05 (Student’s t test). (D) Total number of methylated CpG sites obtained from bisulfite sequencing analysis in HG condition with or without SAM administration. n = 5. (E) miR-200b promoter luciferase activity was measured by calculating the GLUC/SEAP ratio in NG- or HG-exposed HMECs with or without SAM. n = 4, *p < 0.05, F = 10.80 (one-way ANOVA). (F) Endothelial function analysis: matrigel tube length (upper panel), ac-LDL uptake (middle panel), and VE-cadherin expression (lower panel) after SAM treatment in HG condition. n = 5, *p < 0.05 (Student’s t test). (G) Immunofluorescence staining of vWF in SAM-treated HMECs in HG condition. n = 5, *p < 0.001 (Student’s t test). Data represented as the mean ± SD.