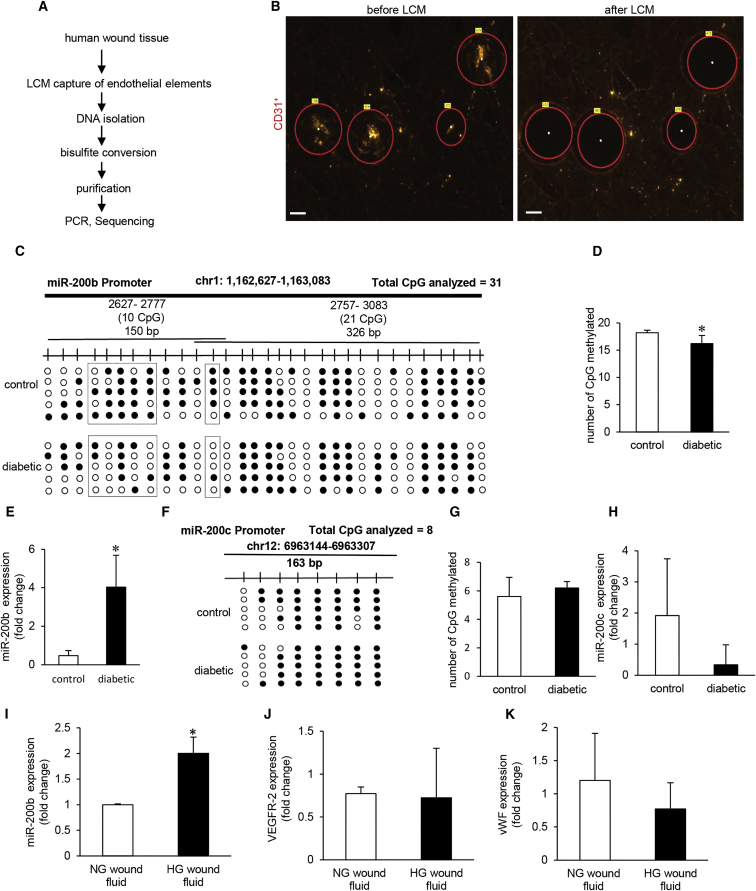
Figure 3.
Promoter Hypomethylation Renders MicroRNA-200b Non-responsive to Injury in Diabetic Wounds
(A) Schematic diagram showing experimental design of miR-200b promoter methylation analysis in human chronic wounds. (B) Representative figure shows the selection of CD31+ tissue elements (red) and their collection before and after the laser capture microdissection (LCM). Scale bar, 150 μm. (C and D) Methylation profile (C) and quantitation of methylated CpG islands in miR-200b promoter (D) in diabetic wounds compared to normoglycemic wounds. N = 5, *p < 0.001 (Student’s t test). (E) qRT-PCR analysis of miR-200b levels in LCM captured endothelial elements from diabetic and non-diabetic wounds. n = 3, *p < 0.05 (Student’s t test). (F) Schematic diagram showing the region of miR-200c promoter analyzed by bisulfite genomic sequencing. Methylation profile of the miR-200c gene promoter in LCM captured endothelial elements from diabetic and non-diabetic wounds with methylated CpGs shown in black. (G) Quantitation of methylated CpG islands in miR-200c promoter in diabetic wounds compared to normoglycemic wounds. n = 5, p = NS (Student’s t test). (H) qRT-PCR analysis of miR-200c expression in diabetic wounds compared to normoglycemic wounds. n = 4, p = NS (Student’s t test). (I) qRT-PCR analysis of miR-200b levels in HMECs after exposure to wound fluid (10%, 4 days). n = 3, *p < 0.05 (Student’s t test). (J and K) qRT-PCR analysis of VEGFR-2 (J) and vWF (K) in HMECs after exposure to wound fluid. n = 3, p = NS (Student’s t test). Data represented as the mean ± SD.