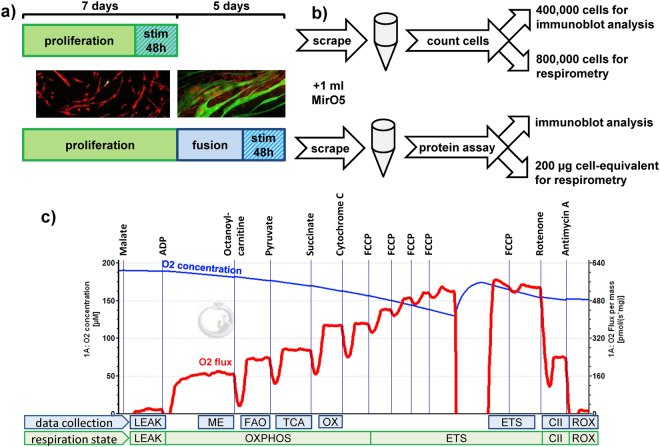
Figure 2.
Workflow for characterisation of mitochondrial function using high resolution respirometry and immunoblot. (a) Myoblasts were seeded on 15 cm dishes and after 80–90% subconfluence differentiated for five days to myotubes. Stimulation with TGFβ1 was performed two days before subconfluence or the last two days of differentiation. (b) Cells were collected by scraping in 1 ml MirO5. Myoblasts were counted and 400,000 cells were collected for protein extraction while 800,000 cells were used for respirometry. For myotubes, 400 µl scraped lysate was pelleted and protein was extracted and quantified. A volume containing 200 µg cellular protein was then used for respirometry. Protein extracts were used for immunoblot analysis. (c) High respiration respirometry of myoblasts and myotubes was carried out using the shown injection order. A representative respiration measurement of myotubes, normalised to mg protein, is shown. Different states of respiration are calculated. The states indicated at “data collection” denote where the measurements reported in this manuscript were obtained. LEAK: respiration corresponding to proton leak. ME: malic-enzyme dependent respiration. FAO: fatty acid oxidation, increase after addition of octanoylcarnitine. TCA: respiration, after addition of all substrates that need to pass through the tricarboxylic acid cycle. OX: oxidative phosphorylation (OXPHOS) after addition of succinate and all TCA substrates. ETS: maximal capacity of the electron transport system after uncoupling. CII: complex II respiration after blocking complex I by rotenone addition. ROX: non-mitochondrial/residual respiration after blocking complex III with antimycin A.