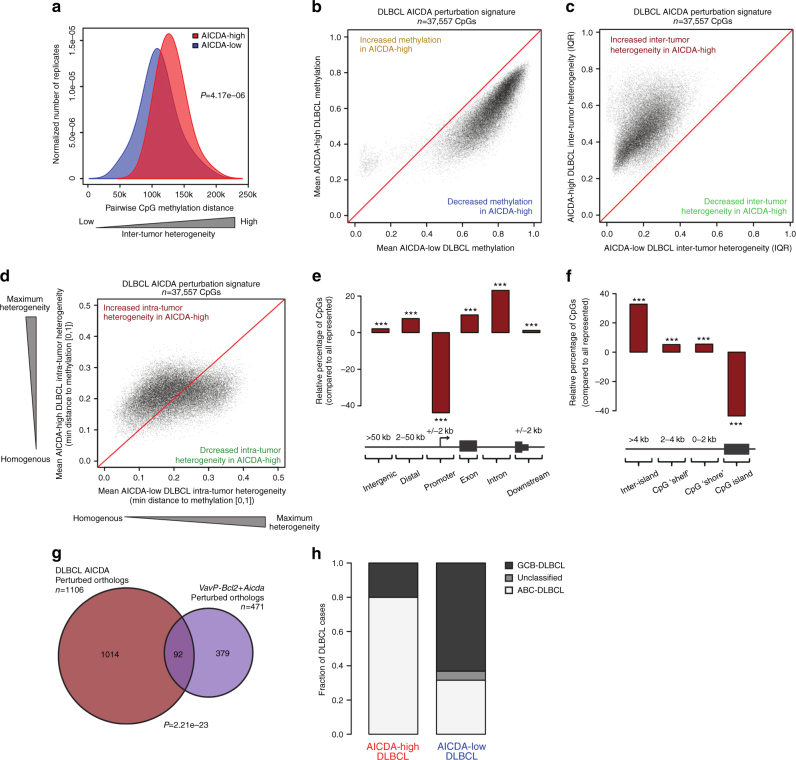
Fig. 4.
High expression of AICDA in DLBCL causes higher DNA methylation heterogeneity and hypomethylation. a Density plot showing the inter-tumor pairwise methylation distance between ERRBS profiles of AICDA-low and AICDA-high DLBCL. AICDA-high DLBCL have greater pairwise distance, indicating increased inter-tumor heterogeneity; two-sided Wilcoxon's signed-rank test. b–d Scatterplots showing shift in mean methylation (b), inter-tumor diversity (c), and intra-tumor heterogeneity (d) of DLBCL AICDA perturbation signature between AICDA-low and AICDA-high DLBCL. e Bar plot showing the distribution of DLBCL AICDA-perturbed CpGs relative to the distribution of all represented CpGs; Fisher’s exact test. f Bar plot showing the relative distribution of DLBCL AICDA-perturbed CpGs within proximity to CpG islands; Fisher’s exact test. g Venn diagram showing the overlap between genes significantly over-representing DLBCL AICDA signature CpGs and murine orthologs over-representing VavP-Bcl2+Aicda signature CpGs; hypergeometric test. h Bar plot showing the relative fraction of DLBCL subtypes within AICDA-high and AICDA-low DLBCL cases (*P < 0.05, **P < 0.01, ***P < 0.001)