Fig. 1.
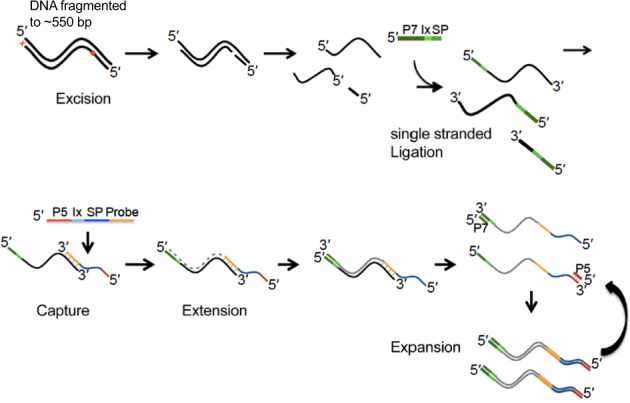
Overview of in-solution OS-Seq process. DNA fragmented to 550 bp is used as the starting material for the OS-Seq assay. Damaged bases are removed by excision only, without implementing a corrective repair step. The DNA is then denatured followed by adapter ligation to single stranded DNA. In-solution capture using primer-probes is performed for ~2 h, followed immediately by extension to complete the library. Finally, the sequence library is expanded by PCR using primers targeting the P7 and P5 regions to generate sufficient quantities of library for sequencing. 5’ and 3’ ends indicated, P7 and P5 indicate regions of adapters and probes, respectively, required for clustering on the Illumina flow cell, or in the “expansion” section, they indicate PCR primers complementary to the P7 and P5 parts of the adapters and probes, respectively. “Ix” stands for index sequence, and “SP” for sequencing primer-binding site
