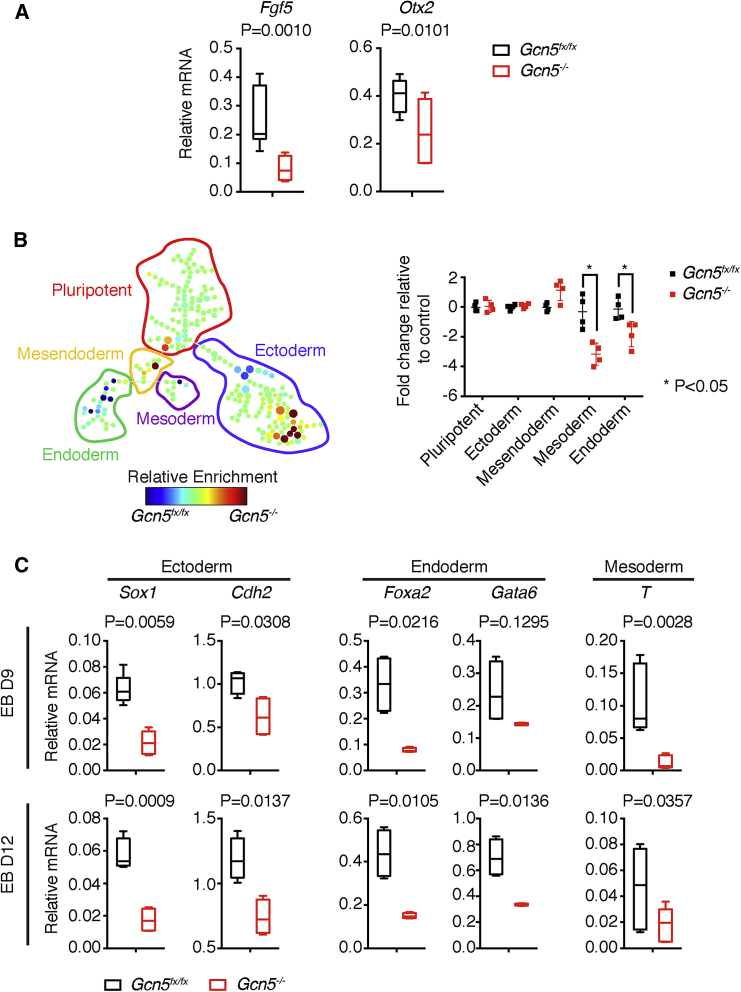
Figure 2.
Compromised Differentiation Potentials of Gcn5 Null EBs
(A) Gene expression analysis by quantitative real-time PCR for epiblast marker genes.
(B) Changes in population composition in Gcn5 null EBs at epiblast stage. Left: SPADE tree plot showing decreased endoderm and mesoderm populations in the Gcn5 null EBs at day 5; Right: quantitation of fold changes in cell numbers of a given population in Gcn5 null EBs relative to control.
(C) Quantitative real-time PCR plots showing decreased expression of marker genes for ectoderm (Sox1, Cdh2), endoderm (Foxa2, Gata6), and mesoderm (T) in late-stage EBs (days 9 and 12).
Data are presented as means ± SD from three (A and C) or four (B) independent experiments.
See also Figures S2 and S3 and Table S1.