Figure 5.
TRIM28 Is Required to Maintain DNA Methylation at Paternal but Not Maternal ICRs in Primed hESCs
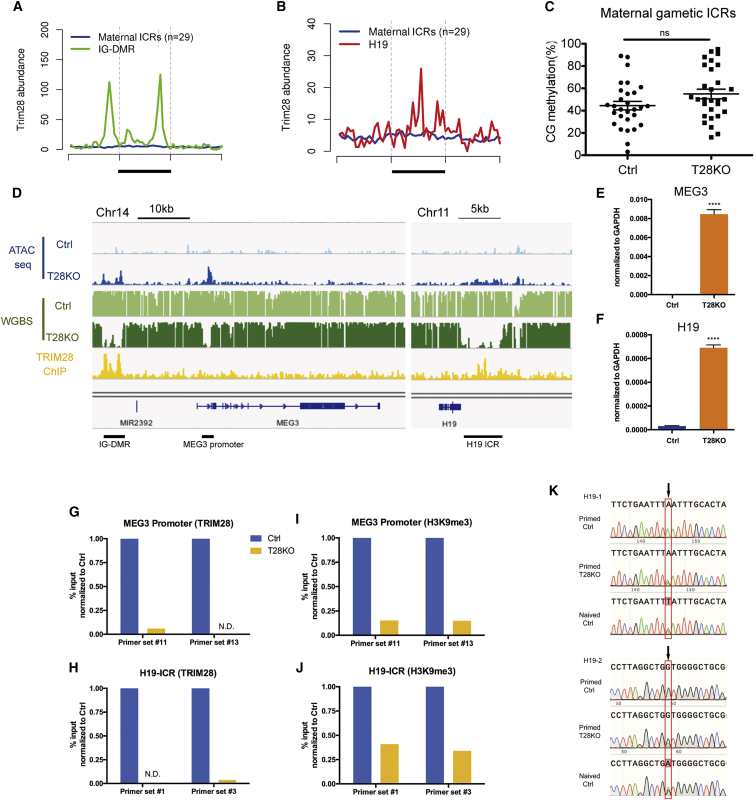
(A and B) TRIM28 binding at the IG-DMR and the H19-ICR relative to the average TRIM28 binding at 29 maternal ICRs as a control. ICR is underlined.
(C) Percent CpG methylation levels at maternal imprinted ICRs in Ctrl and T28KO(U1-9) primed hESCs (n = 29 maternal ICRs), ns, not significant. Error bars represent SD.
(D) Browser view of ATAC-seq (blue), WGBS (green), and TRIM28 ChIP-seq (yellow) at the IG-DMR ICR, the MEG3 promoter, and the H19 ICR.
(E and F) Relative expression using real-time RT-PCR of MEG3 and H19 in T28KO(U1-9) primed hESCs relative to Ctrl. ∗∗∗∗p < 0.0001. Error bars represent SD.
(G–J) ChIP-qPCR of TRIM28 and H3K9me3 at the MEG3 promoter and H19-ICR, in Ctrl and T28KO(U1-9) hESCs.
(K) Sanger sequencing of SNPs in primed Ctrl and T28KO(U1-9) hESCs and naive hESCs as a control for biallelic expression. Arrows indicate two independent H19 SNPs.