Figure 4.
p38α Regulates the Expression of Genes Related to Luminal Cell Specification
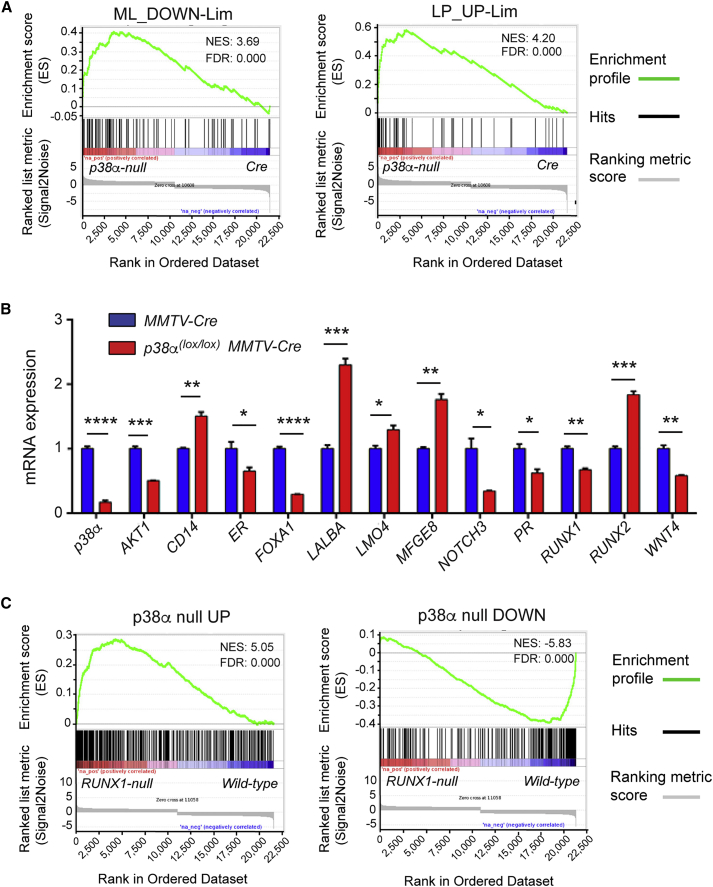
(A) Gene set enrichment analysis (GSEA) plots showing the correlation between the genetic signature of p38α null luminal cells and previously reported signatures of genes downregulated in mature luminal cells (ML_DOWN) or upregulated in luminal progenitor cells (LP_UP) (Lim et al., 2010). Hits mark the position of genes in the published signatures of ML or LP cells according to the fold changes in the p38α null cells from UP (left) to DOWN (right).
(B) Genes known to play a role in luminal cell linage specification and maintenance were analyzed by qRT-PCR in luminal cells isolated from mammary glands of mice with the indicated genotypes (n = 3 independent experiments). Values refer to the expression level of each gene in cells from MMTV-Cre mice, which was given the value of 1. ∗p ≤ 0.05; ∗∗p ≤ 0.005; ∗∗∗p ≤ 0.0005; ∗∗∗∗p ≤ 0.00005.
(C) GSEA plots showing the correlation between genes upregulated (p38α null UP) or downregulated (p38α null DOWN) in p38α null luminal cells and expression fold changes of the genes in RUNX1 null cells from published data. Hits refer to genes differentially expressed in p38α null cells, which are sorted according to the fold changes in the RUNX1 null cells from UP (left) to DOWN (right).
See also Figure S3.