Abstract
The present study aimed to assess the expression of growth arrest-specific 5 (GAS5) and microRNA (miR)-21 in systemic lupus erythematosus (SLE), and attempted to explore their association with clinical features. CD4+ T cells were isolated from peripheral blood of healthy donors and SLE patients by magnetic-activated cell sorting. GAS5 and miR-21 expression levels in cluster of differentiation (CD)4+ T cells were measured by reverse-transcription quantitative polymerase chain reaction. The results revealed that GAS5 and miR-21 levels were significantly elevated in CD4+ T cells of patients with SLE compared with those in control subjects (P<0.05). Regarding clinical features, SLE patients with ulceration had higher GAS5 expression levels in CD4+ T cells than those without ulceration (P<0.05), and the expression of miR-21 was significantly higher in CD4+ T cells of SLE patients with low levels of complement component 3 (C3) than in those with normal levels of complement C3 (P<0.05). In conclusion, GAS5 and miR-21 in CD4+ T cells may serve as potential biomarkers for the diagnosis and monitoring of the progression of SLE.
Keywords: systemic lupus erythematosus, long non-coding RNA, growth arrest-specific 5, microRNA-21
Introduction
Systemic lupus erythematosus (SLE) is a multisystemic, chronic inflammatory autoimmune disease of unknown etiology. It is characterized by various pathogenic autoantibodies and immune complexes as well as damage to multiple organ systems, and the course of SLE is long followed by remission and acute attack. The prevalence varies from 31–70/100,000 individuals in China and from 20–70/100,000 individuals worldwide. It is sex-associated, occurring nine times more often in women than in men, particularly in women of child-bearing age (15–35 years) (1–3). The interaction between certain genetic and environmental factors, including chemical factors, viruses and drugs, damages normal immune tolerance and contributes to SLE. Dysfunctional T cells interact with new antigens constantly, leading to a persistent autoimmune reaction (4,5). Previous studies have confirmed that the abnormal activation and proliferation of self-reactive cluster of differentiation (CD)4+ T lymphocytes have a central role in SLE. CD4+ T cells interact with antigen-specific B cells, to make the latter ones become more effective and produce autoantibodies. In addition, CD4+ T cells produce various cytokines when they are activated, which may engender inflammatory reactions (6,7).
Long non-coding RNAs (lncRNAs) are a class of non-protein-coding RNA with transcripts longer than 200 nucleotides. They have numerous important biological functions through different molecular mechanisms, and are closely associated with a variety of clinical diseases, including tumors, metabolic disease and autoimmune diseases. Growth arrest-specific 5 (GAS5), a non-coding gene that hosts a number of small nucleolar RNAs, has been suggested to have numerous important roles in apoptosis and cell growth inhibition. Previous studies have reported that human GAS5 was upregulated in osteoarthritis (OA) patients (8) and downregulated in certain cancer types, including breast cancer (9), renal cell carcinoma (10) and hepatocellular cancer (11). The GAS5 gene locus in the mouse BXSB strain has been linked to increased susceptibility to SLE (12). GAS5 competes with glucocorticoid (GC) response elements (GRE) by interacting with the DNA binding domain of glucocorticoid receptors (GRs). As GCs are potent immunosuppressants, increased lncRNA GAS5 expression and activity in immune or immune-accessory cells may suppress GC action and contribute to the development of autoimmune diseases (13,14).
MicroRNAs (miRNAs) are endogenous non-coding single-stranded RNAs of ~22 nucleotides in length, which regulate gene expression by targeting mRNA for cleavage or translational repression. miRNAs are involved in diverse biological processes, including cell growth, differentiation, apoptosis and the stability of the immune system. miR-21 was initially known as an ‘oncomiR’, as it was identified to be tightly associated with oncogenesis. Multiple studies have identified that miR-21 is overexpressed in numerous diseases, including breast (15), brain (16), esophageal (17) and gastric cancers (18), cardiovascular diseases (19) and autoimmunity diseases (20). However, to the best of our knowledge, studies relevant to GAS5 in CD4+ T cells of SLE patients are still lacking. The present study aimed to investigate whether the expression levels of GAS5 and miR-21 in CD4+ T cells were abnormal in SLE patients, and the association of their levels with clinical manifestations was assessed in an attempt to identify novel molecular biomarkers involved in the pathogenesis of SLE.
Materials and methods
Patients and healthy controls
A total of 45 SLE patients (41 females, 4 males; age, 34.1±1.2 years; disease duration, 4.2±0.6 years) were recruited from the Rheumatology Department of Yijishan Hospital (Wuhu, China). All patients met the 1982 American College of Rheumatology classification criteria for SLE. SLE activity was assessed using the SLE Disease Activity Index (SLEDAI-2K) (21). Furthermore, 30 control subjects (27 females, 3 males; age, 35.9±1.5 years) were frequency-matched with the patients for age and sex. All participants were from of Han Chinese ethnicity. Clinical information on the patients is listed in Table I. The present study was approved by the Research Ethics Board of Yijishan Hospital Affiliated to Wannan Medical College (Wuhu, China). Written informed consent was obtained from all study participants.
Table I.
Clinical features of patients with SLE.
| Parameter | SLE patients (n=45) | Control (n=30) |
|---|---|---|
| Age (years) | 34.1±1.2 | 35.9±1.5 |
| Sex (n) | ||
| Female | 41 | 27 |
| Male | 4 | 3 |
| Anti-dsDNA (P/N)a | 20/23 | – |
| LN (P/N) | 23/22 | – |
| C3 levela | ||
| <80 mg/dl | 25 | – |
| ≥80 mg/dl | 17 | – |
| Disease duration (years) | 4.2±0.6 | – |
| SLEDAI-2K score | 11.4±1.1 | – |
| Medical therapy | ||
| Prednisone dose ≥30 mg/day | 21 | – |
| Prednisone dose <30 mg/day | 24 | – |
| Immunosuppressants (P/N)b | 19/26 | – |
As certain patients were not examined, the number listed for the feature is less than the total number of patients.
Immunosuppressants included cyclophosphamide, methotrexate, cyclosporine A, chloroquine, leflunomide and mycophenolate mofetil. Values are expressed as n or as the mean ± standard error of the mean. Anti-dsDNA, anti-double-stranded DNA; LN, lupus nephritis; P/N, positive/negative; SLE, systemic lupus erythematosus; DAI, disease activity index.
Isolation of CD4+ T cells from peripheral blood and RNA processing
Peripheral blood samples were obtained from each subject. The samples were collected in tubes containing Heparin sodium. Peripheral blood mononuclear cells (PBMCs) were isolated from anticoagulated whole blood by use of Ficoll density gradient centrifugation. CD4+ T cells were purified from PBMCs by magnetic-activated cell sorting, according to the manufacturer's instructions. PBMCs were successively incubated with fluorescein isothiocyanate (FITC) mouse anti-human CD4 antibody (BD Pharmingen, Franklin Lakes, NJ, USA) and anti-FITC MicroBeads antibody (Miltenyi Biotec, Bergisch Gladbach, Germany). Cell suspension was applied onto a magnetic separation column (Miltenyi Biotec); CD4+ T cells remained in the column and were collected in buffer. The purity rate of the CD4+ T cells (typically 92%) was detected using flow cytometry. Total RNA was then extracted from CD4+ T cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The concentration and purity of RNA were measured by SmartSpec™. Plus spectrophotometry (A260:A280, >1.8) and the integrity of RNA was checked by agarose gel electrophoresis with ethidium bromide staining. The total RNA samples were kept at −80°C prior to use.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
The RT reaction of GAS5 and miR-21 was performed using a Thermo Scientific RevertAid First Strand cDNA Synthesis kit (cat. no. k1622; Thermo Fisher Scientific, Inc.) and a miScript II RT kit (cat. no. 218161; Qiagen, Hilden, Germany). The RT reaction conditions for GAS5 were as follows: Initial incubation at 65°C for 5 min, then at 42°C for 50 min and 70°C for 15 min. The RT conditions for miR-21 were 37°C for 60 min, 95°C for 5 min and a holding step on ice. The total complementary (c)DNA samples were kept at −20°C before use.
PCR amplification of cDNA of GAS5 and miR-21 was performed using the CFX96 real-time system-C1000 thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Real-time PCR of GAS5 and miR-21 was performed in duplicate or triplicate using a QuantiNova SYBR-Green PCR kit (cat. no. 208052), and a miScript SYBR-Green PCR kit (cat. no. 218073) (both from Qiagen), respectively. The following primers were used: GAS5 forward, 5′-AGCTGGAAGTTGAAATGG-3′ and reverse, 5′-CAAGCCGACTCTCCATACC-3′; β-actin forward, 5′-TGACGTGGACATCCGCAAAG-3′ and reverse, 5′-CTGGAAGGTGGACAGCAGGG-3′. The catalogue numbers for the miR-21 and U6 primers were MS00009079 and MS00033740 (both from Qiagen), respectively. The reaction conditions for GAS5 contained an initial heat activation step at 95°C for 5 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec. Conditions for qPCR of miR-21 were as follows: Initial heat activation at 95°C for 15 min, followed by 40 cycles of 95°C for 15 sec, 55°C for 30 sec and 70°C for 30 sec. The dissociation curves of the used primer pairs were generated to confirm a single peak. The mean of the quantification cycle (Cq) was calculated for the reactions. The expression of GAS5 was compared between patients and control subjects by normalizing to β-actin, and miR-21 was normalized to U6. Relative quantification was performed using the 2−ΔΔCq method (22).
Statistical analysis
All statistical analyses were performed using GraphPad Prism version 5.0 software (GraphPad Software, La Jolla, CA, USA). Values are expressed as the mean ± standard error of the mean. Differences in gene expression between two groups were assessed using a Mann-Whitney U test. A two-tailed P<0.05 was considered to indicate a statistically significant difference.
Results
GAS5 and miR-21 expression levels in CD4+ T cells of controls and SLE patients
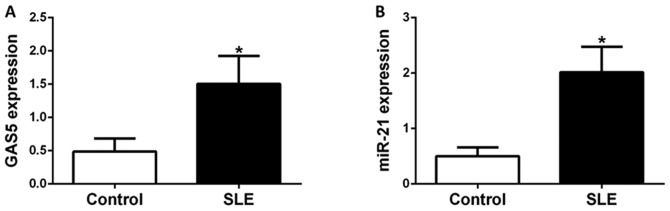
CD4+ T cells obtained from 30 healthy donors and 45 SLE patients were isolated for gene expression analyses. The expression levels of GAS5 and miR-21 in CD4+ T cells were evaluated by RT-qPCR. Patients and control subjects were sex and age-matched. The average disease duration of patients with SLE enrolled in the present study was 4.2 years, with a mean SLEDAI-2K score of 11.4. Anti-double-stranded (ds)DNA, lupus nephritis (LN) and complement C3 levels are important indicators of SLE disease activity and assessed by SLEDAI-2K. In the present study, 20 patients had anti-dsDNA, 23 patients had LN and 25 had low levels of complement C3. GCs and immunosuppressants are the two main types of drug for treating SLE. In the present study, 21 patients were treated with prednisone (dose, ≥30 mg/day) and 19 were treated with immunosuppressants (Table I). GAS5 and miR-21 expression was significantly higher in patients with SLE than in healthy donors (P<0.05). These results indicated that higher expression of GAS5 and miR-21 was specific for SLE and that GAS5 and miR-21 may contribute to the pathogenesis of SLE (Fig. 1).
Figure 1.
GAS5 and miR-21 expression in SLE patients. (A) GAS5 and (B) miR-21 expression were significantly higher in SLE patients than in healthy donors. Values are expressed as the mean ± standard error of the mean. *P<0.05 vs. control. GAS5, growth arrest-specific 5; miR, microRNA.
Association of GAS5 expression in CD4+ T cells and clinical features
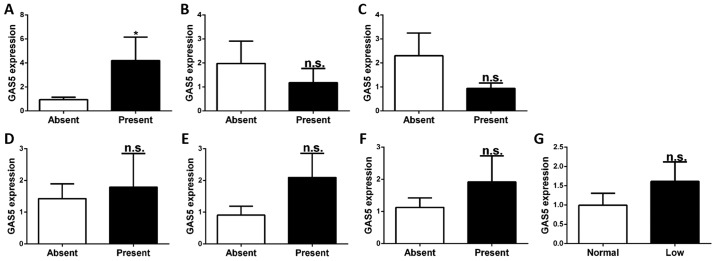
SLE is a systemic autoimmune disease with various clinical features affecting various tissues. Certain clinical features are correlated with disease activity and progression. To investigate whether the expression of GAS5 in CD4+ T cells is associated with these clinical features (nephritis, arthritis, ulceration, pleurisy, rash, anti-dsDNA and complement C3), SLE patients were divided into sets of two groups according to the presence or absence of these respective clinical features. Regarding GAS5 expression in each of these group pair sets, the levels of GAS5 were higher in patients with ulceration than in those without ulceration (P<0.05). However, there were no significant differences in GAS5 expression regarding other clinical features (nephritis, arthritis, pleurisy, rash, anti-dsDNA and complement C3) (P>0.05; Fig. 2).
Figure 2.
Expression of GAS5 in patients with different clinical features. (A) GAS5 was significant higher in patients with ulceration. However, no significant differences were found for (B) nephritis, (C) arthritis, (D) pleurisy, (E) rash, (F) anti-double stranded DNA and (G) complement component 3. Values are expressed as the mean ± standard error of the mean. *P<0.05 vs. control. n.s., no significance; GAS5, growth arrest-specific 5.
Association of miR-21 expression in CD4+ T cells and clinical features
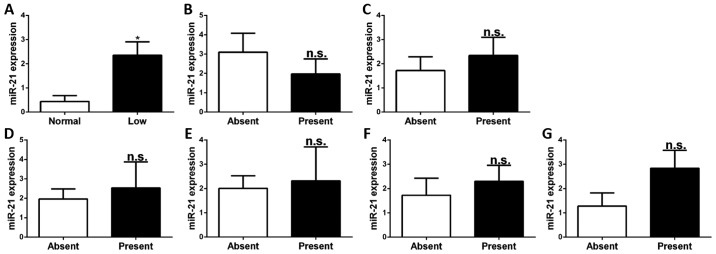
To investigate whether the expression of miR-21 in CD4+ T cells is associated with the abovementioned clinical features, the relative expression levels of miR-21 in CD4+ T cells of SLE patients stratified by the presence or absence of these clinical features were compared. It was identified that the levels of miR-21 in CD4+ T cells of patients with low levels of complement C3 were higher than in those with normal levels of complement C3 (P<0.05). There was no association between miR-21 expression and any of the other clinical features, namely nephritis, arthritis, ulceration, pleurisy, rash and anti-dsDNA (P>0.05; Fig. 3).
Figure 3.
Expression of miR-21 in different clinical features. (A) miR-21 was significant higher in patients with low level of complement component 3. And no significant difference was found in (B) nephritis, (C) arthritis, (D) ulceration, (E) pleurisy, (F) rash and (G) anti-double stranded DNA. Values are expressed as the mean ± standard error of the mean. *P<0.05 vs. control. n.s., no significance; miR, microRNA.
Discussion
SLE is an autoimmune disease with a complex and unpredictable course. It is known that GCs are still the first-line drugs for SLE. However, chronic high-dose hormone therapy may give rise to adverse events and GC resistance. Previous studies have demonstrated that aberrant expression and binding of the GR may be associated with GC resistance in SLE patients, and that it may be considered as a biomarker to personalize therapy (23,24). Little is known about the influence of GAS5 on the susceptibility for SLE and its prevention. The present study detected for lncRNA GAS5 and miR-21, and investigated the association between their expression levels and specific clinical features of SLE. The results revealed that GAS5 and miR-21 levels were significantly elevated in patients with SLE compared with those in control subjects. The results regarding miR-21 were consistent with those of a previous study (20). The results of the present study indicated that GAS5 and miR-21 expressed in CD4+ T cells were specific for SLE and may contribute to its pathogenesis. Among the clinical features of SLE, ulceration and complement C3 levels are two indicators of disease activity according to the SLE Disease Activity Index (SLEDAI-2K). GAS5 levels in CD4+ T cells were identified to be higher in patients with ulceration than in those without ulceration, and miR-21 levels in CD4+ T cells were higher in patients with low levels of complement C3 than in those with normal levels of complement C3.
To the best of our knowledge, the present study was the first to report an association of GAS5 and miR-21 in CD4+ T cells with ulceration and complement C3 levels, respectively, in patients with SLE. Although the detailed mechanisms remain to be fully elucidated, GAS5 and miR-21 levels in CD4+ T cells may be two key indicators of disease activity in patients with SLE. The levels of GAS5 in CD4+ T cells had an increasing trend in patients with pleurisy, rash, anti-dsDNA and low complement C3, and the levels of miR-21 in CD4+ T cells had an increasing trend in patients with arthritis, ulceration, pleurisy, rash and anti-dsDNA; however, there was no significance. All of these results suggested that GAS5 and miR-21 levels in CD4+ T cells may be useful for predicting the progression of SLE. It has been reported that lncRNA GAS5 was negatively regulated by miR-21 in breast tumors, hepatocellular carcinoma and osteoarthritis. GAS5 was also capable of suppressing miR-21 through an lncRNA/miRNA interaction, implying a feedback loop between GAS5 and miR-21 (8,25–27). However, in the present study, GAS5 as well as miR-21 were identified to be upregulated in the CD4+ T cells of SLE patients, which may be associated with different types of diseases and cells (8,28,29). In the CD4+ T cells of SLE patients, the function of GAS5 may be primarily dependent on glucocorticoids signaling pathways (13). Future study is required to clarify the association between GAS5 and miR-21 in SLE.
GAS5 has been reported to be closely associated with various human diseases (8,28,30). GAS5 functions as a potential tumor suppressor and is downregulated in several types of cancer (28). GAS5 is also involved in the regulation of mammalian cell apoptosis and cell population growth (31–33). The downregulation of GAS5 protects T cell lines as well as untransformed human T-lymphocytes (34). GAS5, a 5′-terminal oligopyrimidine RNA, whose translation is specifically controlled by the mammalian target of rapamycin pathway, is required for the inhibition of human T cell proliferation by rapamycin and its analogues (35). The function of GAS5 is dependent on its direct association with the GR protein; GAS5 binds to the GR through mimicking GRE and acts as a decoy GRE, thus blocking the upregulation of gene transcription. GR target genes are involved in apoptosis suppression, such as cellular inhibitor of apoptosis 2 and serum/GC-regulated kinase 1, and inhibit the cell-death executioners caspase-3, −7 and −9. As GCs are powerful immunosuppressants, most of the known biological actions of GCs are mediated by the GR. The present study identified that the GAS5 levels in SLE patients were higher than those in the control group, indicating that increased lncRNA GAS5 expression in CD4+ T cells suppressed GC action and contributed to the development of SLE (13,14).
Previous studies have suggested that miR-21 functions as an anti-apoptotic and pro-survival factor in numerous cell types, and miR-21 was the only miRNA upregulated in all of the tumor types analyzed (29,36). Programmed cell death protein 4 (PDCD4), novel tumor suppressor gene, is a direct target gene of miR-21. A previous study supported that the miR-21/PDCD4 controlled pathway has a central role in SLE (37). Aberrant DNA methylation was also reported to be involved in the progression of SLE. The present study found that miR-21 was overexpressed in CD4+ T cells from patients with SLE, which promoted cell hypomethylation by repressing DNA methyltransferase 1 expression, induced the overexpression of autoimmune-associated methylation-sensitive genes and mediated the pathogenesis of SLE (20).
In conclusion, the present study revealed that GAS5 and miR-21 levels in CD4+ T cells were significantly elevated in patients with SLE compared with those in control subjects. Regarding the clinical features of SLE, the expression of GAS5 and miR-21 in CD4+ T cells was associated with ulceration and low complement C3, respectively. GAS5 and miR-21 in CD4+ T cells may serve as two potential biomarkers for the diagnosis and prediction of the progression of SLE.
Acknowledgements
The present study was supported by the key research grant of Wannan Medical College (grant no. WK20142F04).
References
- 1.Squatrito D, Emmi G, Silvestri E, Ciucciarelli L, D'Elios MM, Prisco D, Emmi L. Pathogenesis and potential therapeutic targets in systemic lupus erythematosus: From bench to bedside. Auto Immun Highlights. 2014;5:33–45. doi: 10.1007/s13317-014-0058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Y, Zhang F, Ma J, Zhang X, Wu L, Qu B, Xia S, Chen S, Tang Y, Shen N. Association of large intergenic noncoding RNA expression with disease activity and organ damage in systemic lupus erythematosus. Arthritis Res Ther. 2015;17:131. doi: 10.1186/s13075-015-0632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Wu Z, Zhang S, Chen S, Li P, Li J, Cao C, Liu B, Zhang F, Li Y. Genetic variants of IkappaB kinase β (IKBKB) and polymerase β (POLB) were not associated with systemic lupus erythematosus risk in a Chinese Han population. PLoS One. 2015;10:e0132556. doi: 10.1371/journal.pone.0132556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosalka J, Jakiela B, Musial J. Changes of memory B- and T-cell subsets in lupus nephritis patients. Folia Histochem Cytobiol. 2016;54:32–41. doi: 10.5603/FHC.a2016.0005. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, Liao J, Zhao M, Wu H, Yung S, Chan TM, Yoshimura A, Lu Q. Increased expression of TLR2 in CD4(+) T cells from SLE patients enhances immune reactivity and promotes IL-17 expression through histone modifications. Eur J Immunol. 2015;45:2683–2693. doi: 10.1002/eji.201445219. [DOI] [PubMed] [Google Scholar]
- 6.Rottman JB, Willis CR. Mouse models of systemic lupus erythematosus reveal a complex pathogenesis. Vet Pathol. 2010;47:664–676. doi: 10.1177/0300985810370005. [DOI] [PubMed] [Google Scholar]
- 7.Bakshi J, Ismajli M, Rahman A. New therapeutic avenues in SLE. Best Pract Res Clin Rheumatol. 2015;29:794–809. doi: 10.1016/j.berh.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Song J, Ahn C, Chun CH, Jin EJ. A long non-coding RNA, GAS5, plays a critical role in the regulation of miR-21 during osteoarthritis. J Orthop Res. 2014;32:1628–1635. doi: 10.1002/jor.22718. [DOI] [PubMed] [Google Scholar]
- 9.Pickard MR, Williams GT. The hormone response element mimic sequence of GAS5 lncRNA is sufficient to induce apoptosis in breast cancer cells. Oncotarget. 2016;7:10104–10116. doi: 10.18632/oncotarget.7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiao HP, Gao WS, Huo JX, Yang ZS. Long non-coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pac J Cancer Prev. 2013;14:1077–1082. doi: 10.7314/APJCP.2013.14.2.1077. [DOI] [PubMed] [Google Scholar]
- 11.Chang L, Li C, Lan T, Wu L, Yuan Y, Liu Q, Liu Z. Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Mol Med Rep. 2016;13:1541–1550. doi: 10.3892/mmr.2015.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haywood ME, Rose SJ, Horswell S, Lees MJ, Fu G, Walport MJ, Morley BJ. Overlapping BXSB congenic intervals, in combination with microarray gene expression, reveal novel lupus candidate genes. Genes Immun. 2006;7:250–263. doi: 10.1038/sj.gene.6364294. [DOI] [PubMed] [Google Scholar]
- 13.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Y, Cai Q, Huang Y, Li S, Yang H, Sun L, Chen K, Wang Y. MicroRNA-21 as a potential diagnostic biomarker for breast cancer patients: A pooled analysis of individual studies. Oncotarget. 2016;7:34498–34506. doi: 10.18632/oncotarget.9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu K, Lin T, Pang Q, Liu T, Wang Z, Tai M, Meng F, Zhang J, Wan Y, Mao P, et al. Extracellular miRNA-21 as a novel biomarker in glioma: Evidence from meta-analysis, clinical validation and experimental investigations. Oncotarget. 2016;7:33994–34010. doi: 10.18632/oncotarget.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu YR, Qi HJ, Deng DF, Luo YY, Yang SL. MicroRNA-21 promotes cell proliferation, migration, and resistance to apoptosis through PTEN/PI3K/AKT signaling pathway in esophageal cancer. Tumour Biol. 2016;37:12061–12070. doi: 10.1007/s13277-016-5074-2. [DOI] [PubMed] [Google Scholar]
- 18.Sekar D, Krishnan R, Thirugnanasambantham K, Rajasekaran B, Islam VI, Sekar P. Significance of microRNA 21 in gastric cancer. Clin Res Hepatol Gastroenterol. 2016;40:538–545. doi: 10.1016/j.clinre.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Jazbutyte V, Thum T. MicroRNA-21: From cancer to cardiovascular disease. Current Drug Targets. 2010;11:926–935. doi: 10.2174/138945010791591403. [DOI] [PubMed] [Google Scholar]
- 20.Pan W, Zhu S, Yuan M, Cui H, Wang L, Luo X, Li J, Zhou H, Tang Y, Shen N. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J Immunol. 2010;184:6773–6781. doi: 10.4049/jimmunol.0904060. [DOI] [PubMed] [Google Scholar]
- 21.Gladman DD, Ibañez D, Urowitz MB. Systemic lupus erythematosus disease activity index 2000. J Rheumatol. 2002;29:288–291. [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Lucafò M, Bravin V, Tommasini A, Martelossi S, Rabach I, Ventura A, Decorti G, De Iudicibus S. Differential expression of GAS5 in rapamycin-induced reversion of glucocorticoid resistance. Clin Exp Pharmacol Physiol. 2016;43:602–605. doi: 10.1111/1440-1681.12572. [DOI] [PubMed] [Google Scholar]
- 24.Du J, Li M, Zhang D, Zhu X, Zhang W, Gu W, Feng Y, Zhai X, Ling C. Flow cytometry analysis of glucocorticoid receptor expression and binding in steroid-sensitive and steroid-resistant patients with systemic lupus erythematosus. Arthritis Res Ther. 2009;11:R108. doi: 10.1186/ar2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu L, Ye H, Huang G, Luo F, Liu Y, Liu Y, Yang X, Shen J, Liu Q, Zhang J. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumour Biol. 2016;37:2691–2702. doi: 10.1007/s13277-015-4111-x. [DOI] [PubMed] [Google Scholar]
- 26.Pickard MR, Williams GT. Molecular and cellular mechanisms of action of tumour suppressor GAS5 lncRNA. Genes (Besel) 2015;6:484–499. doi: 10.3390/genes6030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, Wu F, Mo YY. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma C, Shi X, Zhu Q, Li Q, Liu Y, Yao Y, Song Y. The growth arrest-specific transcript 5 (GAS5): A pivotal tumor suppressor long noncoding RNA in human cancers. Tumour Biol. 2016;37:1437–1444. doi: 10.1007/s13277-015-4521-9. [DOI] [PubMed] [Google Scholar]
- 29.Gao Y, Dai M, Liu H, He W, Lin S, Yuan T, Chen H, Dai S. Diagnostic value of circulating miR-21: An update meta-analysis in various cancers and validation in endometrial cancer. Oncotarget. 2016;7:68894–68908. doi: 10.18632/oncotarget.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter G, Miladinovic B, Patel AA, Deland L, Mastorides S, Patel NA. Circulating long noncoding RNA GAS5 levels are correlated to prevalence of type 2 diabetes mellitus. BBA Clin. 2015;4:102–107. doi: 10.1016/j.bbacli.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu X, Fang Y, Wang Z, Xie J, Zhan Q, Deng X, Chen H, Jin J, Peng C, Li H, Shen B. Downregulation of gas5 increases pancreatic cancer cell proliferation by regulating CDK6. Cell Tissue Res. 2013;354:891–896. doi: 10.1007/s00441-013-1711-x. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z, Wang W, Jiang J, Bao E, Xu D, Zeng Y, Tao L, Qiu J. Downregulation of GAS5 promotes bladder cancer cell proliferation, partly by regulating CDK6. PLoS One. 2013;8:e73991. doi: 10.1371/journal.pone.0073991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mourtada-Maarabouni M, Williams GT. Growth arrest on inhibition of nonsense-mediated decay is mediated by noncoding RNA GAS5. Biomed Res Int. 2013;2013:358015. doi: 10.1155/2013/358015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu G, Lou Z, Gupta M. The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS One. 2014;9:e107016. doi: 10.1371/journal.pone.0107016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams GT, Mourtada-Maarabouni M, Farzaneh F. A critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytes. Biochem Soc Trans. 2011;39:482–486. doi: 10.1042/BST0390482. [DOI] [PubMed] [Google Scholar]
- 36.Bertoli G, Cava C, Castiglioni I. MicroRNAs: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pratheeshkumar P, Son YO, Divya SP, Wang L, Turcios L, Roy RV, Hitron JA, Kim D, Dai J, Asha P, et al. Quercetin inhibits Cr(VI)-induced malignant cell transformation by targeting miR-21-PDCD4 signaling pathway. Oncotarget. 2016;8:52118–52131. doi: 10.18632/oncotarget.10130. [DOI] [PMC free article] [PubMed] [Google Scholar]