Fig. 2.
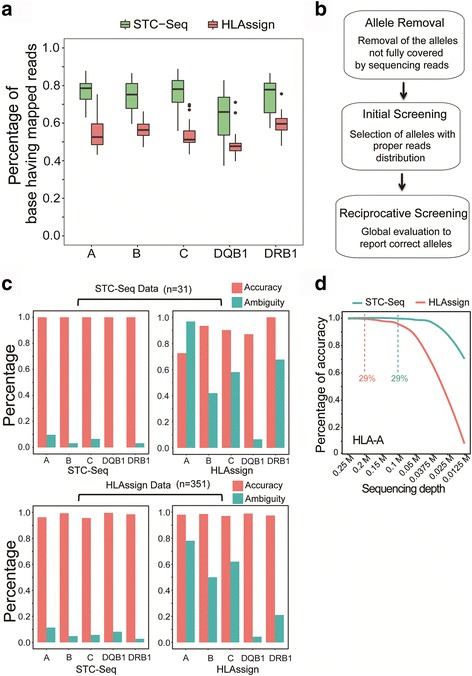
Comparison of HLA-typing accuracy between STC-Seq and HLAssign. a Comparison of the percentages of bases having mapped reads between STC-seq and HLAssign. The HLA-A/B/C/DQB1/DRB1 core exons (2,3,4) were compared. Only the positions of the first bases of the mapped reads were considered. b Overview of the STC-Seq analysis pipeline. c Comparison of typing accuracy of HLA-A/B/C/DQB1/DRB1 between STC-seq and HLAssign. d Comparison of the typing accuracy of HLA-A between 31 STC-Seq datasets and 31 HLAssign datasets at different sequencing depths using STC-seq analysis pipeline. The dashed lines indicate the sequencing depths of STC-seq and HLAssign datasets at a threshold with 29% exon bases having mapped reads, below which the typing accuracy drops significantly
