Fig. 1.
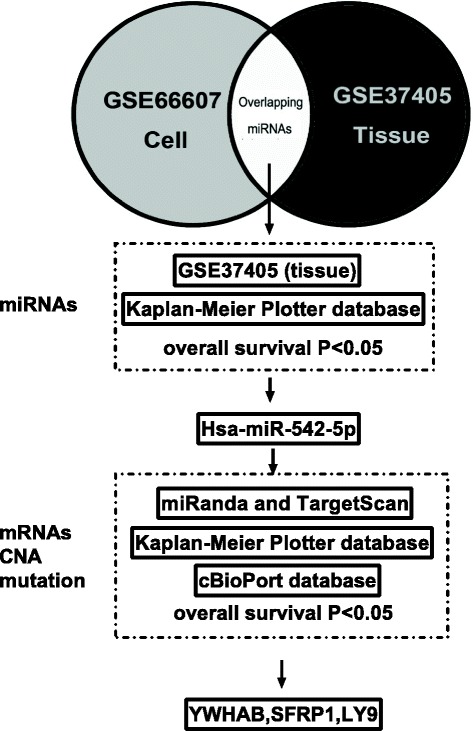
The multi-step strategy used in this study. First, significantly changed miRNAs in GSE66607 (cells for miRNA in MCF-7/TamR vs MCF-7) and GSE37405 (tissues for breast cancer tissues) were identified. Second, GSE37405 and Kaplan-Meier Plotter Database (KMPD) for miRpower of breast cancer as the validated set were used to screen overlapped miRNAs in step 1 that related to overall survival. Third, for the target mRNA genes, KMPD, and Gene Expression Profiling Interactive Analysis (GEPIA), were used to reveal the relationship between target genes and overall survival
