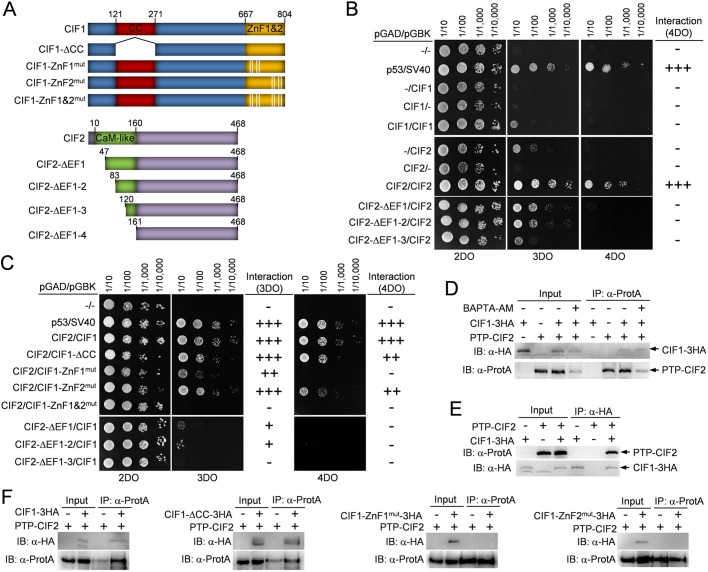
Fig. 2.
Identification of the structural motifs involved in CIF1-CIF2 interaction. (A) Schematic illustration of wild-type and mutant CIF1 and CIF2. CIF1-ΔCC, CIF1 coiled-coil motif deletion mutant; CIF1-ZnF1mut, CIF1 zinc-finger 1 mutant generated by mutating Cys693, Cys696, His708 and Cys712 to alanine residue; CIF1-ZnF2mut, CIF1 zinc-finger 2 mutant generated by mutating Cys769, Cys772, His784 and Cys788 to alanine; CIF2-ΔEF1, CIF2 EF-hand motif 1 deletion mutant by deleting aa 1–46; CIF2-ΔEF1-2, CIF2 EF-hand motif 1–2 deletion mutant by deleting aa 1–82; CIF2-ΔEF1-3, CIF2 EF-hand motifs 1–3 deletion mutant by deleting aa 1–119; CIF2-ΔEF1-4, CIF2 EF-hand motifs 1–4 deletion mutant by deleting aa 1–160. (B) Yeast two-hybrid assays to test the potential self-interaction of CIF1 and CIF2. 2DO medium (SD −Leu −Trp) selects the presence of both the pGAD and pGBK plasmids. 3DO medium (SD –Leu –Trp −His) detects protein–protein interaction under medium-stringency conditions, and 4DO medium (SD –Leu –Trp –His –Ade) detects protein–protein interaction under high-stringency conditions. Note that due to the self-activation of CIF2 on 3DO medium, protein–protein interactions were only scored based on the 4DO medium. (C) Yeast two-hybrid assays to test the potential interaction between CIF1 and CIF2 and the requirement of structural motifs for CIF1–CIF2 interaction. (D) Co-immunoprecipitation to test the interaction between PTP-tagged CIF2 and 3HA-tagged CIF1 in the absence or presence of the calcium chelator BAPTA-AM. Immunoprecipitation was carried out by pulling down PTP–CIF2 with IgG beads, and the co-immunoprecipitated CIF1–3HA and PTP–CIF2 were detected by anti-HA monoclonal and anti-Protein-A (α-ProtA) polyclonal antibodies, respectively. Cells expressing PTP–CIF2 alone and CIF1–3HA alone were included as negative controls. (E) Reciprocal immunoprecipitation to test the interaction between CIF1–3HA and PTP–CIF2. Immunoprecipitation was carried out by pulling down CIF1–3HA with anti-HA agarose beads, and co-immunoprecipitated PTP–CIF2 and CIF1–3HA were detected by anti-Protein-A and anti-HA antibodies. Cells expressing CIF1–3HA alone and PTP–CIF2 alone are included as negative controls. (F) Co-immunoprecipitation to test the potential interaction between CIF2 and CIF1 mutants. Wild-type and mutant CIF1 proteins were tagged with a triple HA epitope and ectopically expressed in the CIF1-3′UTR RNAi cell line, and CIF2 was tagged with a PTP epitope from its endogenous locus in the CIF1-3′UTR RNAi cells expressing wild-type and mutant CIF1 proteins. PTP–CIF2 was immunoprecipitated by IgG beads, and the immunoprecipitated proteins were immunoblotted with anti-HA antibody to detect 3HA-tagged wild-type and mutant CIF1 and with anti-Protein-A to detect PTP–CIF2.