ABSTRACT
The Slit–Robo GTPase-activating proteins (srGAPs) were first identified as potential Slit–Robo effectors that influence growth cone guidance. Given their N-terminal F-BAR, central GAP and C-terminal SH3 domains, srGAPs have the potential to affect membrane dynamics, Rho family GTPase activity and other binding partners. Recent research has clarified how srGAP family members act in distinct ways at the cell membrane, and has expanded our understanding of the roles of srGAPs in neuronal and non-neuronal cells. Gene duplication of the human-specific paralog of srGAP2 has resulted in srGAP2 family proteins that may have increased the density of dendritic spines and promoted neoteny of the human brain during crucial periods of human evolution, underscoring the importance of srGAPs in the unique sculpting of the human brain. Importantly, srGAPs also play roles outside of the nervous system, including during contact inhibition of cell movement and in establishing and maintaining cell adhesions in epithelia. Changes in srGAP expression may contribute to neurodevelopmental disorders, cancer metastasis and inflammation. As discussed in this Review, much remains to be discovered about how this interesting family of proteins functions in a diverse set of processes in metazoans and the functional roles srGAPs play in human disease.
KEY WORDS: Slit–Robo GAP, Brain evolution, srGAP2, C. elegans, Cell–cell adhesion, Contact inhibition
Summary: A review of the functions of Slit–Robo GTPase-activating proteins (srGAPs), which are highly conserved proteins that interact with the plasma membrane, regulate Rho family GTPases, and function in a wide range of cell biological processes.
Introduction
As cells migrate, change shape and modulate cell–cell contacts, they must dynamically modulate the curvature of their phospholipid membranes and recruit different protein complexes to microdomains that exhibit differences in curvature and/or lipid composition. The Bin-Amphiphysin-Rvs (BAR) domain is one of several protein motifs that allows cells to interact with phospholipid membranes with a range of curvatures (for in-depth reviews of membrane curvature-generation, see Jarsch et al., 2016; McMahon and Boucrot, 2015). BAR domains homodimerize and are localized to membranes through electrostatic interactions between positively charged residues on the BAR domain and negatively charged lipids (Frost et al., 2008; Mim and Unger, 2012) (Fig. 1A).
Fig. 1.
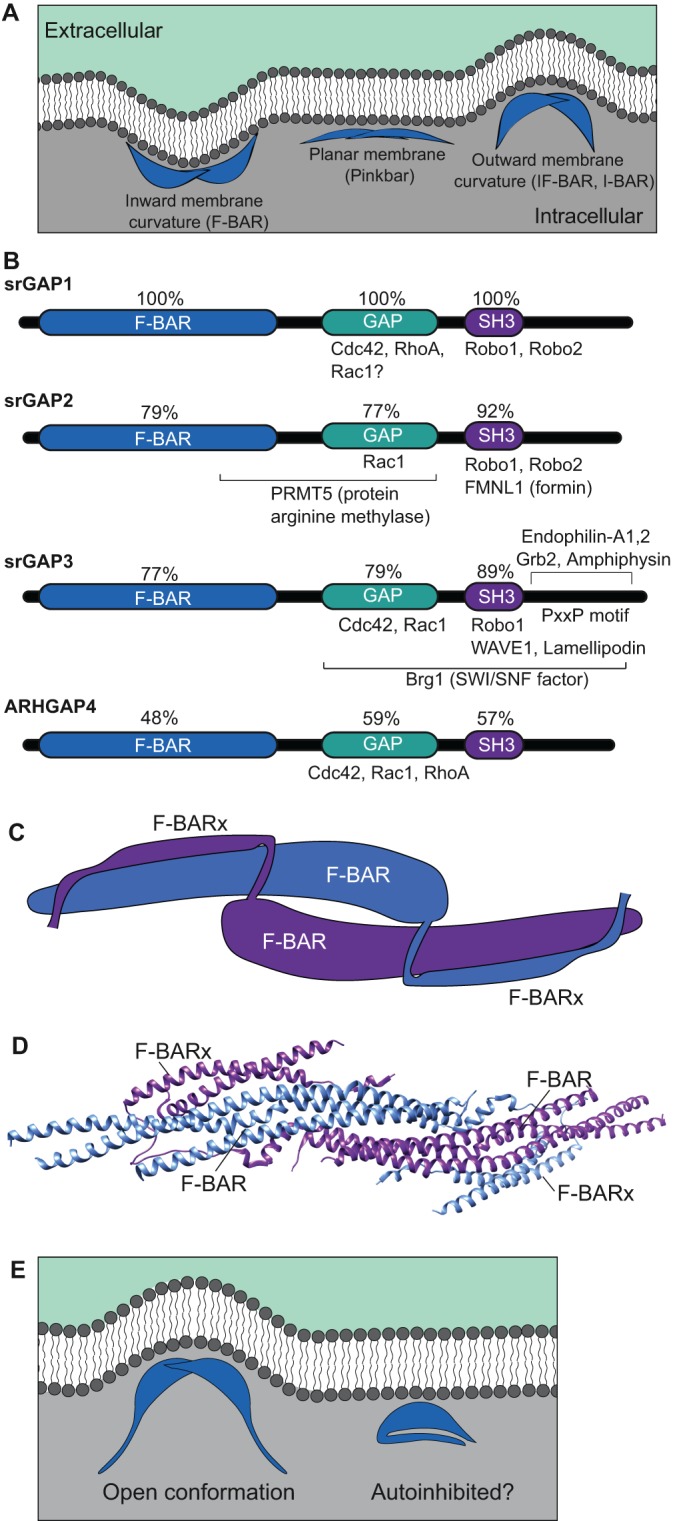
srGAPs are BAR domain proteins with characteristic functional domains. (A) BAR domains (blue) allow proteins to dimerize and interact with specific phospholipids (dark gray) at the membrane. FCH-Bin, Amphiphysin, Rvs (F-BAR) domains interface with membranes through their concave surfaces. Inverse BAR (I-BAR) and inverse F-BAR (IF-BAR) domains associate with membranes through their convex surfaces, generally interfacing with evaginations. Pinkbar associates with non-curved planar membranes through its convex surface. (B) Domain structure of srGAPs. Vertebrate srGAPs have three highly conserved domains: the lipid-binding F-BAR domain, a GAP domain that serves to inactivate Rho GTPase family members, and the SH3 domain, which is important for multiple protein-protein interactions. Regions mediating known binding interactions are indicated for each family member; see main text for references. (C) Diagrammatic representation of two srGAP2 monomers within an inferred homodimer, based on a rendering of PDB 5I6J, as originally reported in Sporny et al. (2017). The F-BAR and extended F-BAR domains (F-BARx) interact to stabilize the homodimer. (D) Structure of the presumed homodimer based on crystallographic data (PDB 5I6J; rendered using UCSF Chimera). (E) Possible model for autoinhibition of the F-BAR domain. When the F-BAR domains of two srGAP proteins homodimerize, membrane evaginations are generated (left).
In this Review, we focus on a class of BAR domain-containing proteins that has generated significant interest in both the neurodevelopmental and evolutionary fields, the Slit–Robo GTPase-activating proteins (srGAPs; Fig. 1B). srGAPs were originally named because their C-terminal SH3 domain binds the intercellular domain of the Roundabout (Robo) family receptors, whose canonical ligands are members of the Slit family of secreted proteins (Wong et al., 2001). srGAPs are prevalent in neural tissues (Bacon et al., 2009; Chen et al., 2012b; Waltereit et al., 2008; Wong et al., 2001), and their loss can lead to severe congenital neurological disabilities in humans (Chen et al., 2012a; Ellery et al., 2014; Endris et al., 2002; Gunnarsson and Foyn Bruun, 2010; Peltekova et al., 2012; Riess et al., 2012; Rincic et al., 2016; Shuib et al., 2009).
There are four srGAPs in vertebrates: srGAP1, also known as Rho GTPase-activating protein 13 (ARHGAP13); srGAP2, also known as formin-binding protein 2 (FNBP2) or ARHGAP14; srGAP3, also known as Wiskott–Aldrich syndrome protein family Verprolin-homologous protein (WAVE)-associated RacGAP Protein (WRP) or mental disorder-associated GAP (MEGAP); and srGAP4, commonly referred to as ARHGAP4, and also known as C1 or p115 Rho GAP. srGAP1, srGAP2 and srGAP3 are 60–80% identical to one another, whereas ARHGAP4 is less well conserved, with only 51% identity to srGAP3 (BLAST; https://blast.ncbi.nlm.nih.gov/; Fig. 1B). Through their centrally located GTPase-activating protein (GAP) domains, the srGAPs regulate the activity of RhoGTPase family members, in turn modulating cytoskeletal dynamics during a wide variety of processes, both within and outside of the nervous system. In addition to conferring the ability to bind Robo receptors, the C-terminal SH3 domain of srGAPs is critical for their interactions with proteins involved in regulating actin architecture, cell migration and cell adhesion, such as lamellipodin (also known as RAPH1) (Endris et al., 2011) and Wave1 (also known as WASF1) (Soderling et al., 2002). Here, we examine recent findings that clarify how srGAPs interact with membranes and with their effectors, and how srGAPs may influence diverse cellular events in neuronal and non-neuronal tissues.
srGAPs display varying phospholipid-binding specificities
srGAPs contain a specialized type of BAR domain, known as an inverse F-BAR (IF-BAR), which induces evaginations in cell culture (Carlson et al., 2011; Endris et al., 2011; Guerrier et al., 2009; Ma et al., 2013; Sporny et al., 2017; Wang et al., 2014; Zaidel-Bar et al., 2010); the presence of a unique F-BAR extension (F-BARx) may help produce this membrane curvature (Guerrier et al., 2009; Sporny et al., 2016) (Fig. 1C,D). Like other BAR domain-containing proteins, srGAPs can exist in an autoinhibited state with regards to lipid binding (Rao et al., 2010), which may be facilitated by interactions between the SH3 domain and the F-BAR domain, thereby preventing the protein from effectively inducing membrane bends (Guerrier et al., 2009; Guez-Haddad et al., 2015) (Fig. 1C), similar to how Drosophila Nervous Wreck and human BIN1 act (Chen et al., 2014; Kelley et al., 2015; Rao et al., 2010; Stanishneva-Konovalova et al., 2016; Vázquez et al., 2013; Wu and Baumgart, 2014). The N- and C-termini of SRGP-1, the single srGAP in Caenorhabditis elegans, directly physically interact, supporting this hypothesis (Zaidel-Bar et al., 2010). Furthermore, the F-BAR domains of srGAP1, srGAP2 and srGAP3 (Coutinho-Budd et al., 2012) and SRGP-1 (Zaidel-Bar et al., 2010) all induce longer filopodia than the full-length proteins. Such results are consistent with C-terminus-induced autoinhibition of membrane bending, which is mediated by the N-terminus; alternatively, the C-terminus of srGAPs could have additional roles in restricting membrane evagination that are independent of direct binding to the N-terminus.
srGAP2 and srGAP3 interact with lipids through positively charged residues on one surface of their F-BAR domains (Carlson et al., 2011; Sporny et al., 2017). Because of the similarity in curvature induced by srGAP1, srGAP2 and srGAP3 and their high level of sequence identity (Coutinho-Budd et al., 2012), it is assumed that the general mechanism by which they interact with membranes is similar. Nevertheless, the three proteins do exhibit differences in lipid-binding specificity and effects on membrane curvature. srGAP2 is recruited by lipids other than phosphatidylinositol (4,5)-bisphosphate, which is a key lipid in recruiting both srGAP1 and srGAP3. Additionally, srGAP2 more potently facilitates filopodium formation (in terms of both number and length) than srGAP1 and srGAP3. Although, in general, srGAPs promote membrane evagination, intriguingly, in some contexts the F-BAR domain of srGAP1 appears to restrict membrane bending (Coutinho-Budd et al., 2012). Moreover, srGAP1 to 3 have differing localizations along filopodia, with srGAP2 uniquely localizing to tips of extending filopodia. How the three proteins work together to promote the formation of membrane bends in some areas and to restrict them elsewhere remains to be determined.
Perhaps the most crucial question about the interactions of srGAPs with the plasma membrane relates to precisely what functional role the F-BAR domain plays in vivo. Do F-BAR domains bend membranes, sense their curvature, or both? Answering this question is surprisingly difficult. That overexpression of srGAPs and especially their F-BAR domains leads to membrane evaginations is clear (Guerrier et al., 2009; Zaidel-Bar et al., 2010) and indicates that srGAPs can bend membrane outward. But is production of additional curved membrane surface via srGAPs necessary? Structure–function experiments in C. elegans in srgp-1 loss-of-function backgrounds indicates that the F-BAR domain is necessary to keep SRGP-1 in close proximity to the plasma membrane (Zaidel-Bar et al., 2010; our observations). These findings also suggest that induction of curvature is a normal function of SRGP-1, since the leading edge of migrating epidermal cells displays reduced curvature when SRGP-1 is depleted (Zaidel-Bar et al., 2010). Such studies are, however, far from definitive; moreover, these studies do not cleanly separate curvature recognition from curvature induction. Doing so would require introducing mutations into srGAPs that prevent membrane bending without preventing binding to curved membranes. No such mutations are currently known. Perhaps, at present, on balance, it is best to say that srGAPs likely create curvature, in addition to whatever other functions they may play in recognizing membranes that have been curved via other means.
srGAPs show varying specificities for Rho family GTPases
As is true for their lipid-binding specificities, various srGAPs appear to have both unique and overlapping GAP specificities: srGAP1 inactivates Cdc42 and RhoA (Wong et al., 2001), srGAP2 specifically stimulates the GTPase activity of Rac1 (Guerrier et al., 2009; Ma et al., 2013; Mason et al., 2011), and srGAP3 inactivates both Cdc42 and Rac1 (Soderling et al., 2002; Waltereit et al., 2012; Yang et al., 2006). Whether srGAP1 also inactivates Rac1 is controversial (Ma et al., 2013; Wong et al., 2001; Yamazaki et al., 2013), although expression of a constitutively active Rac1 increases membrane recruitment of srGAP1, supporting an interaction (Yamazaki et al., 2013). Finally, ARHGAP4 has a less-specific RhoGAP activity, as it stimulates the GTPase activity of Rac1, Cdc42 and RhoA in rats (Foletta et al., 2002), although another study found that ARHGAP4 only inactivates RhoA in mice (Christerson et al., 2002). SRGP-1 in C. elegans has specificity mainly for CED-10/Rac (Neukomm et al., 2011), which may represent an evolutionarily basic function of srGAPs.
The srGAP C-terminus acts as a scaffold for protein interaction
Although the F-BAR domain and GAP activity are both crucial for srGAP function, they are not the only important domains in the srGAPs. Through their C-terminal SH3 domains, srGAPs interact with other proteins that contribute to cell migration (for schematics, see Fig. 1B). srGAP3, for example, localizes to focal adhesions and interacts with lamellipodin through its SH3 domain (Endris et al., 2011). Lamellipodin, which is negatively regulated by srGAP3, is important for modulating actin dynamics at the leading edge of cells (Chang et al., 2006; Hansen and Mullins, 2015; Krause et al., 2004; Quinn et al., 2008). Similarly, the SH3 domain of srGAP2 interacts with the formin FMNL1 and both directly and indirectly inhibits its actin-severing capabilities (Mason et al., 2011). Furthermore, ARHGAP4 interacts with MAP/ERK kinase kinase 1 (MEKK1; also known as MAP3K1) (Christerson et al., 2002). At least for srGAP2, regions flanking the SH3 domain appear to modulate high-affinity ligand binding at the SH3 site itself (Guez-Haddad et al., 2015).
The most C-terminal portion of the srGAPs is poorly conserved, both among the vertebrate srGAPs (Wong et al., 2001; Wuertenberger and Groemping, 2015) and when compared to SRGP-1 in C. elegans (Zaidel-Bar et al., 2010). C-terminal truncations of ARHGAP4 fail to regulate axon outgrowth (Vogt et al., 2007), suggesting the C-terminal regions are important for function. Although the functional importance of the interaction is currently unknown, the C-terminus of srGAP3 is responsible for its interaction with the BAR domain proteins endophilin-A1 and endophilin-A2, as well as with amphiphysin, all of which are involved in synaptic vesicle endocytosis (Wuertenberger and Groemping, 2015). Other potential srGAP interactors include the protein arginine methyltransferase PRMT5 (arginine methylated srGAP2; Guo and Bao, 2010) and the SWI/SNF chromatin remodeler Brg1 (srGAP3; Dai et al., 2014; see Fig. 1B). Clearly, further elucidation of srGAP interactors will enhance our understanding of the role of the different srGAPs as protein scaffolds.
One potentially useful avenue for elucidating evolutionarily fundamental aspects of the function of the C-terminus of srGAPs is the single srGAP family member in C. elegans, SRGP-1. Although it has well-conserved F-BAR and GAP modules, SRGP-1 lacks an SH3 domain (Zaidel-Bar et al., 2010). Given the known promiscuity of SH3 domains as protein scaffolds, the fact that SRGP-1 lacks this domain may mean that it has fewer C-terminal-binding partners than human srGAPs, potentially simplifying the analysis of SH3-independent C-terminal-binding partners. Of particular interest is whether SRGP-1 retains the ability to bind the C. elegans Robo homolog SAX-3 given that the Slit–Robo system in C. elegans, in many respects, functions in a manner similar to that in vertebrates (Hao et al., 2001; Zallen et al., 1998).
srGAPs modulate neurite outgrowth and spinogenesis
Given the interactions srGAPs engage in through their N-terminal F-BAR domain (membrane lipids), their GAP domain (Rho family GTPases) and their C-terminus (e.g. actin-binding proteins), manipulation of srGAP expression might be expected to affect cell migration. Indeed, increased expression of srGAPs most often leads to a decrease in cell migration (Guerrier et al., 2009; Vogt et al., 2007; Yamazaki et al., 2013; Yang et al., 2006). Filopodium formation, as well as neurite initiation and outgrowth, are promoted by overexpression of srGAP1 (Ma et al., 2013) or srGAP2 (Guerrier et al., 2009; Ma et al., 2013); the ability of srGAP2 to bend membranes is required for this effect (Guerrier et al., 2009).
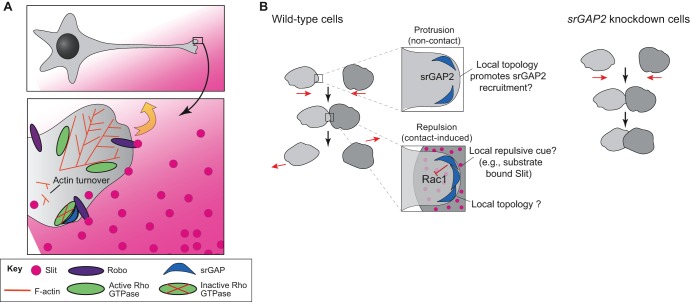
srGAPs have most often been proposed to play roles in axon guidance, downstream of the protein Slit. Slit typically acts as a repellent and is assumed to be a diffusible ligand, although it has also been proposed to act as a signal displayed on the surface of cells (Fritz et al., 2015) through heparan sulfate proteoglycans (HSPGs) (Ypsilanti et al., 2010). In response to binding to the Slit ligand, the CC3 domain of the receptors Robo1 and Robo2 interacts with the SH3 domain of srGAPs (Mason et al., 2011; Wong et al., 2001). The ability of srGAPs to bind Robo receptors, combined with their GAP activity towards Rho family GTPases, suggests that srGAPs could be involved in the spatial regulation of Rho GTPase activity in migrating cells. Recruitment of srGAPs provides an appealing mechanism whereby growth cone repulsion could occur: subcellular recruitment of srGAPs could inactivate Rho GTPases through their GAP activity, thereby reducing local actin polymerization asymmetrically, leading to subsequent turning of the growth cone away from regions of higher Slit concentration (Fig. 2A). Consistent with this model, Slit binding to Robo1 increases the interaction of srGAP1 with Cdc42 (Wong et al., 2001).
Fig. 2.
Role of vertebrate srGAPs in cell repulsion. (A) srGAPs as effectors of Slit–Robo signaling in neurons. The boxed region in the neuron is enlarged below. srGAP (blue) can bind to Robo (purple) receptor when Robo is bound to its ligand Slit (pink). In turn, the srGAPs locally inactive RhoGTPases (green). Because of the asymmetric localization of active RhoGTPases, actin polymerizes asymmetrically, leading to movement away from the Slit cue (yellow arrow). This pathway operates in neuronal cells (Wong et al., 2001), leukocytes (Sherchan et al., 2016; Ye et al., 2010) and fibroblasts (Fritz et al., 2015). The Slit guidance cue can be distributed as a diffusible gradient, as shown here in pink, or may be displayed on the surface of neighboring cells via HSPGs. (B) srGAPs as potential ‘adhesion buffers’ during contact inhibition of locomotion. Red arrows in the central portion of the panel indicate the direction of cell migration. Upon collision, wild-type fibroblasts in culture (left) undergo contact-mediated repulsion and migrate away from one another. Prior to collision (top enlargement), srGAP2 may respond to or modulate membrane topology to promote leading-edge dynamics. After collision (bottom enlargement), additional srGAP2 may be recruited to sites of cell–cell contact, leading to locally decreased Rac1 activity, which promotes repulsion. One proposed mechanism for presenting repellant is HSPG-immobilized Slit on or near the cell surface (Fritz et al., 2015). In contrast, in srGAP2-knockdown cells (right) there is no transduction of repulsive cues to Rac1 inhibition, and cells remain in prolonged contact after collision.
Despite the appeal of models in which srGAPs act to transduce repulsive signals downstream of Slit and Robo, however, clear functional evidence for a direct regulation of growth cone motility by srGAPs (e.g. based on loss-of-function analysis), is surprisingly meager. In the original report that described srGAP1, overexpression of a dominant-negative srGAP1 construct in the forebrain blocked repulsion of migratory cells from the anterior subventricular zone (SVZa) of the forebrain in an explant system (Wong et al., 2001). In srGAP3-knockout mice, loss of srGAP3 results in the abnormal migration of neuroblasts from the ventricular region into the corpus callosum (Kim et al., 2012). srGAP3 and ARHGAP4 reduce cell migration and protrusive activity, in a GAP-dependent manner, when they are overexpressed in cell culture (Vogt et al., 2007; Yang et al., 2006). srGAP3 overexpression in mice reduces neurite outgrowth in a manner dependent on its GAP domain, similar to what occurs upon expression of a dominant-negative form of its target Rac1 (Soderling et al., 2002). These studies are consistent with roles for srGAPs in mediating neuronal migration in response to Slit, but they do not conclusively show that srGAPs function as the essential link between the receipt of Slit repulsive cues by Robo and downregulation of Rho GTPase function. Such a demonstration would require more incisive epistasis testing using loss-of-function mutants and manipulation and/or assessment of Rho GTPase activity.
Although downregulation of Cdc42 or Rac1 may explain much of the inhibitory activity of srGAPs on neuronal outgrowth, the specific molecular mechanisms by which srGAPs negatively affect migration are likely to be more complex than solely through simple direct regulation of Rho GTPases. The propensity of srGAPs to promote membrane bending through their F-BAR domains may also affect the motility of the growth cone, at least in the case of srGAP overexpression; excessive branching of leading processes could lead to decreased cell migration, presumably owing to increased competition among supernumerary protrusions (Guerrier et al., 2009). Further complicating the picture is the cross-regulation of Rho family GTPases. For example, it has been suggested that because srGAP2 and srGAP1 both locally inactivate Rac1, the subsequent activation of RhoA could result in increased filopodium formation through the formin mDia (Guerrier et al., 2009; Yamazaki et al., 2013) (a pathway reviewed in Mellor, 2010).
In addition to their effects on growth cones, srGAPs affect other aspects of neuronal function. In vivo, srGAP3 may be particularly important for the formation of dendritic spines, small extensions orthogonal to the shaft of dendrites that receive inputs from excitatory axons (Kessels and Qualmann, 2015; Lei et al., 2016; Rochefort and Konnerth, 2012). Wave1 mutant mice, in which the Wave1 lesion leads to reduced binding between Wave1 and srGAP3, display spine defects (Soderling et al., 2007). Mice heterozygous or homozygous for a putative null conditional srGAP3 mutation likewise have defects in spine formation (Carlson et al., 2011). srGAP3 homozygous null mice have significantly fewer spines than wild-type mice, and even heterozygous mice have fewer mature mushroom-shaped spines. Similarly, mice heterozygous or homozygous for a mutation that mimics a human srGAP3 deletion have a significantly increased spine length (Waltereit et al., 2012). Another set of studies did not find differences in spine density and morphology frequencies between srGAP3 mutant and wild-type mice, but this may be due to differences in the nature of the lesion (Bertram et al., 2016; Waltereit et al., 2012) or the genetic backgrounds used (Kim et al., 2012; Waltereit et al., 2012).
srGAPs are expressed in different neuronal cell types
In addition to their differential effects at the subcellular level, srGAPs show broad differences in their expression in various neuronal cell types. In situ hybridization shows that srGAP1–3 and ARHGAP4 are expressed throughout the developing central nervous system and portions of the peripheral nervous system in mice and rats, such as in dorsal root ganglia (Bacon et al., 2009; Chen et al., 2012b; Christerson et al., 2002; Foletta et al., 2002; Waltereit et al., 2008; Wong et al., 2001). srGAP2 and srGAP3 are present in zones of proliferation and differentiation in the developing brain, and therefore may be involved in neural differentiation. srGAP2 is expressed in the ventricular and subventricular zones in humans, regions in which neural progenitors divide to produce postmitotic neurons (Charrier et al., 2012). srGAP3 is expressed in rat neural stem cells and/or neural progenitor cells in the subventricular zone. srGAP3 also localizes to the neocortex throughout development and adulthood (Waltereit et al., 2008). Loss of srGAP3 leads to decreased viability and differentiation of these cells, while srGAP2 expression appears to decrease normally during cell differentiation (Jiao et al., 2016; Lu et al., 2013; Ma et al., 2013). srGAP3 is one of only two genes downregulated in ventricular zone progenitors mutant for the key neuronal differentiation transcription factor neurogenin, suggesting that srGAP3 expression may be important during specification of these cells (Mattar et al., 2004). In contrast, srGAP3 appears to inhibit differentiation of Neuro2A cells in culture; this negative effect is dependent on its GAP domain, as the loss of srGAP3 could be rescued by a constitutively active Rac1, but not by a GAP-mutated srGAP3 (Chen et al., 2011; Gao et al., 2016). How GAP activity could negatively regulate neuronal differentiation is unclear.
Given their purported roles in neuronal function, significant attention has focused on srGAPs in the clinic. Chromosomal abnormalities associated with srGAP genes are associated with human neurological disorders and numerous studies in mice support a functional role for srGAPs in the etiology of neurological disorders (see Box 1).
Box 1. srGAPs in neurological disease.
There is tantalizing evidence that srGAPs are crucial to proper neurodevelopment (Ramakers, 2002). The first hint that srGAPs may be playing a role in disease came with the discovery of 3p-syndrome. Individuals with this syndrome have a wide array of symptoms, including intellectual deficits, ptosis (drooping of the upper eyelid) and microcephaly (Schwyzer et al., 1987; Verjaal and De Nef, 1978). Attention has focused on loss of srGAP3 (previously called MEGAP) within the chromosome arm deleted in 3p-patients (Endris et al., 2002; Riess et al., 2012). Patients with the opposite defect, partial trisomy 3p syndrome, in which a small chromosomal region containing srGAP3 is duplicated, display intellectual disability and psychomotor retardation (Bittel et al., 2006; Natera-de Benito et al., 2014). Microdeletion of a chromosomal region that includes srGAP2 in humans may lead to reduced gyrification of the brain (Rincic et al., 2016). Rare copy number variations in an srGAP2 paralog are found in individuals with autism spectrum disorders (Dennis et al., 2012; Nuttle et al., 2013), and a mutation within srGAP2 has been associated with early infantile epileptic encephalopathy (Saitsu et al., 2012). While suggestive, further studies will be required to show that deletion of srGAP genes are the definitive causative agents in these disorders. Studies in mice reinforce these analyses of human patients. Mice deficient for srGAP3 or its interactors have multiple neural impairments, including decreased social behavior and anxiety (Soderling et al., 2003; Waltereit et al., 2012), schizophrenia- and autism-like symptoms (Bertram et al., 2016; Waltereit et al., 2012), hydrocephalus (Bertram et al., 2016; Kim et al., 2012; Koschützke et al., 2015; Waltereit et al., 2012), ciliary defects (Koschützke et al., 2015), and issues with learning and memory (Carlson et al., 2011; Soderling et al., 2007, 2003; Waltereit et al., 2012).
The paralog srGAP2C has a unique role in human brain evolution
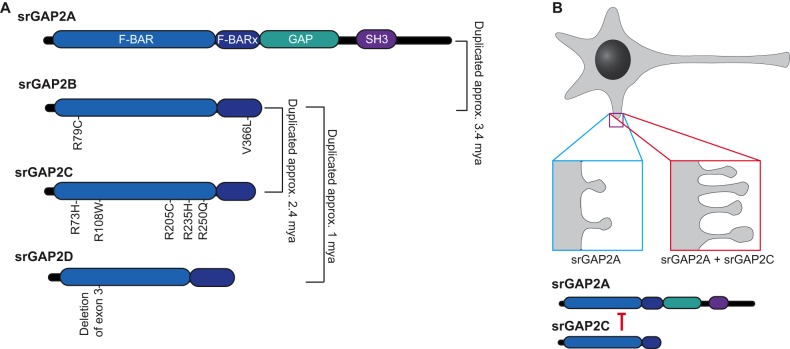
Given their apparent roles in neuronal development and function, one might expect that changes in expression of srGAPs might be correlated with the evolution of the human brain. One piece of evidence for such an evolutionary role arose from improved annotation of the srGAP2 locus, which showed that there were multiple paralogs of srGAP2 on chromosome I, termed srGAP2A, srGAP2B and srGAP2C (Dennis et al., 2012) (Fig. 3A). Two gene duplication events correlate with key milestones in hominin evolution. A gene duplication event ∼3.5 million years ago created a first srGAP2 paralog, srGAP2B. Although the timing of this event is somewhat controversial (Almecija et al., 2015; Skinner et al., 2015b), this duplication occurred at approximately the same time that Australopithecus spp. may have begun using stone tools (Dennis et al., 2012; Skinner et al., 2015a; Thompson et al., 2015). Later, and less controversially, a second paralog, srGAP2C, arose at about the time that the earliest members of the genus Homo were using tools and at a time when significant expansion of the neocortex began (Dennis et al., 2012; Huang et al., 1995). These correlations between gene duplication events and elaboration of brain structures in the Homo lineage provide an exciting path for future evolutionary studies.
Fig. 3.
Evolutionary history of srGAP2 paralogs. (A) srGAP2A, the ancestral form of srGAP2, contains an F-BAR domain, an extended F-BAR domain (F-BARx), a GAP domain and an SH3 domain. srGAP2A is an essential gene. srGAP2B arose as a partial duplication of srGAP2A ∼3.4 million years ago (mya), producing a protein that only contains the F-BAR and F-BARx domains. srGAP2B, in turn, duplicated ∼2.4 mya to produce srGAP2C and again ∼1 mya to produce srGAP2D. After these duplications, srGAP2B accumulated two nonsynonymous mutations (R79C, V366L), while srGAP2C accumulated five crucial nonsynonymous mutations (R73H, R108W, R205C, R235H, R250Q) that are important for antagonism of srGAP2A (Sporny et al., 2017). srGAP2D, likely a nonessential pseudogene, lacks the third exon, which normally encodes part of the F-BAR domain. (B) Non-human mammals express only the equivalent of human srGAP2A, and exhibit dendritic spines that are shorter and less numerous than in humans (blue box). In humans or in rodents that express exogenous srGAP2A and srGAP2C, srGAP2C inhibits srGAP2A, causing the dendritic spine necks to be longer, and the spines themselves to become more densely packed (red box).
How might the srGAP2C paralog have promoted brain evolution? srGAP2C is a truncated paralog of srGAP2A that only contains a partial F-BAR domain (Fig. 3A); it is present in all modern human populations, as well as in Neanderthal and Denisovan remains (Ho et al., 2017). srGAP2C interacts with lipids but is incapable of inducing filopodia, unlike the ancestral srGAP2A (Charrier et al., 2012). The truncated srGAP2C can, however, bind to srGAP2A to form a heterodimer; in this case, srGAP2C binding inhibits the ability of srGAP2A to induce membrane curvature (Charrier et al., 2012; Sporny et al., 2017) and to bind the CC3 domain of Robo1, although its GAP activity remains intact (Sporny et al., 2017) (Fig. 3B). While the CC3 domain of Robo1 may not be the sole region of the C-terminus of Robo1 that interacts with srGAPs, this result does suggest that srGAP2A and srGAP2C heterodimerization can affect recruitment of srGAPs to Robo receptors. Moreover, this result suggests that a further five amino acid changes that are specific to srGAP2C increase its ability to heterodimerize with srGAP2A compared with other srGAP2 paralogs (Sporny et al., 2017). This strong interaction between srGAP2A and srGAP2C, in turn, leads to co-regulation of spine density, timing of spine maturation and spine morphology, which are all keys to the neoteny seen in human brains (Charrier et al., 2012) (Fig. 3B). Furthermore, srGAP2C is able to affect density and maturation of both inhibitory and excitatory synapses by inhibiting multiple domains of srGAP2A (Fossati et al., 2016; Sporny et al., 2017). srGAP2C is uniquely restricted in terms of copy number variation throughout human populations (Dennis et al., 2012), suggesting that there is a strict selection for correct srGAP2C gene dosage. Consistent with this idea, duplications of chromosomal segments that include srGAP2C, which would in turn increase srGAP2C-mediated antagonism of srGAP2A, are associated with intellectual disability and autism spectrum disorders (Dennis et al., 2012).
In conclusion, srGAPs have been implicated in the functions of numerous neuronal cell types in vertebrates, including humans, from growth cones and dendritic spines to larger-scale organization of the brain and the density of synaptic connections associated with emergence of Homo sapiens. The exciting challenge for the future will be to correlate specific biochemical functions of srGAP homo- and hetero-dimers with these larger-scale effects on neuronal tissues.
srGAPs have roles beyond the nervous system
Thus far, the focus of this Review has been on the key roles of srGAPs in the peripheral and central nervous system, including in brain architecture. Several recent studies, however, have demonstrated that srGAPs have more general roles in metazoans. One role involves contact inhibition of locomotion (CIL), a classical behavior of cultured cells in which cells reduce protrusive activity when they collide with other cells (Abercrombie and Heaysman, 1953, 1954). Knockdown of srGAP2 in fibroblasts in vitro leads to a failure of such cell–cell repulsion (Fritz et al., 2015). In this system, srGAP2 spatiotemporally regulates Rac activation downstream of Slit–Robo signals, thereby limiting the extension phase of cell–cell contacts and resulting in a failure of cell repulsion. In this sense, srGAP2 may act as a contact-dependent ‘protrusive activity buffer’ to limit protrusions at sites of cell contact (Fig. 2B).
The in vivo relevance of CIL is not entirely clear, but the above result suggests that srGAPs may have roles at sites of cell–cell contact in the intact tissues of multicellular organisms. Support for this possibility comes from studies in the nematode C. elegans, which has proven to be a convenient model organism to study the role of srGAPs outside of the nervous system. Because SRGP-1 is the sole srGAP homolog in C. elegans, there are no issues of potential functional redundancy. SRGP-1 is ∼32% identical to srGAPs (BLAST; https://blast.ncbi.nlm.nih.gov/), although it lacks the C-terminal SH3 domain of the vertebrate srGAPs (Fig. 4A). SRGP-1 is expressed in neuronal cells, but, significantly, it is also expressed broadly in epithelial tissues. Like other srGAPs, SRGP-1 localizes to curved membranes and induces long membrane evaginations when overexpressed (Zaidel-Bar et al., 2010). Loss of SRGP-1 leads to a delay in the completion of ventral enclosure of the epidermis, which normally spreads to cover underlying cells, predominantly neuroblasts (Fig. 4B). In addition, ventral neuroblasts occasionally fail to seal a transient cleft (termed the ventral cleft) in sensitized genetic backgrounds in which the function of the cadherin–catenin complex is impaired, suggesting SRGP-1 normally aids the establishment of cadherin-dependent cell–cell adhesions during morphogenesis (Zaidel-Bar et al., 2010). How SRGP-1 modulates cell–cell adhesion is unclear. Migrating ventral epidermal cells in srgp-1 mutants have smoother membranes than wild-type embryos. Defects in epithelial sealing events may therefore be due to a decrease in the plasma membrane surface area that is available for junction formation at the leading edge (Fig. 4B). In addition to its role in morphogenesis, SRGP-1 negatively regulates cell corpse engulfment, apparently through its CED-10/RacGAP activity (Neukomm et al., 2011, 2014). Although the SRGP-1 RacGAP domain appears to be less important than its F-BAR domain during epithelial sheet sealing (Zaidel-Bar et al., 2010), it is possible that downregulation of CED-10/Rac contributes to successful epithelial sheet sealing, as downregulation of Rac has been shown to be important for stabilizing cadherin-dependent adhesions (Braga, 2016; Ratheesh et al., 2013; Yamada and Nelson, 2007).
Fig. 4.
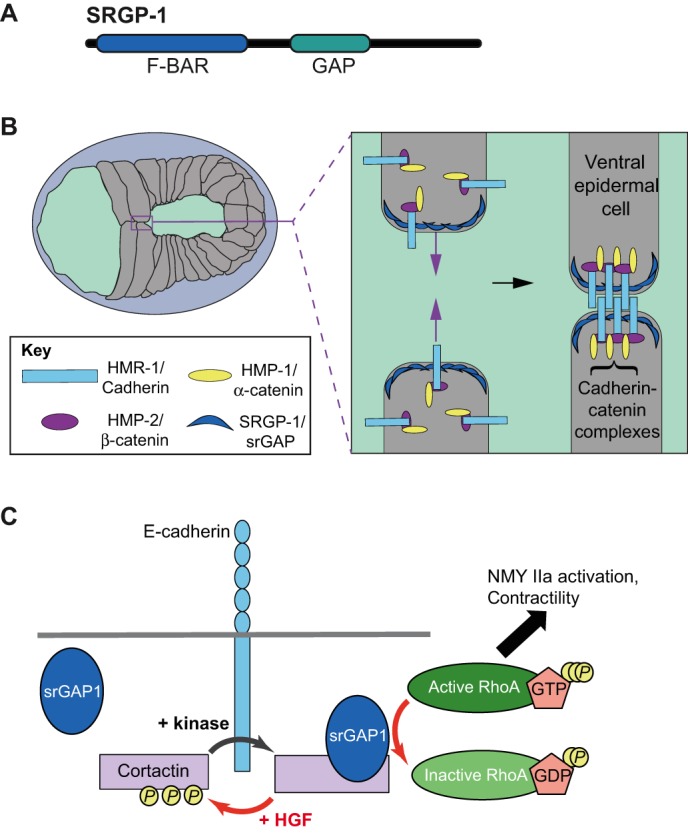
Potential roles of srGAPs in cadherin-based adhesion. (A) Domain map of C. elegans SRGP-1, which lacks the SH3 domain found in vertebrate srGAPs. (B) Schematic of an enclosing C. elegans embryo. The boxed area is expanded on the right. Because epidermal cells make contact at the ventral midline and build nascent junctions, here, SRGP-1 (dark blue crescents) may increase surface area at the leading edge to facilitate efficiency of junction formation. HMR-1/cadherin (light blue box), HMP-2/β-catenin (purple ellipse) and HMP-1/α-catenin (yellow ellipse) form nascent junctions between opposing cells in the enclosing ventral epidermis. (C) Model for HGF-dependent regulation of Rho activity at adherens junctions through cortactin and srGAP1 in Caco2 cells. Tyrosine-phosphorylated cortactin (yellow P) does not associated with srGAP1. Steady-state signaling at junctions may activate a kinase that maintains most cortactin in a phosphorylated state. HGF leads to desphosphorylation of cortactin, allowing recruitment of srGAP1. srGAP1 in turn decreases Rho activity via its GAP domain, thereby destabilizing cadherin-dependent cell–cell adhesion.
Studies of SRGP-1 implicate srGAPs in enhancing nascent adherens junction formation. Recent work in Caco2 cells, however, supports the opposite effect for vertebrate srGAP1 (Liang et al., 2017). Knockdown of the scaffolding protein cortactin leads to reduced Rho activity at E-cadherin-containing cell–cell junctions, weakening adhesion. Rho activity cannot be restored by a mutant cortactin carrying non-phosphorylatable mutations at three key tyrosine residues (3YF cortactin). Interestingly, based on co-immunoprecipitation and mass spectrometry assays, more srGAP1 associates with 3YF cortactin than with wild-type protein; conversely, in vitro phosphorylation of cortactin decreases its association with srGAP1. srGAP1 localizes to adherens junctions in a cortactin-dependent manner, and knockdown of srGAP1 leads to a reduction in RhoA activity at junctions (Liang et al., 2017). Taken together, these results suggest that tyrosine-dephosphorylated cortactin recruits srGAP1 to junctions, where it negatively regulates RhoA and thus weakens adherens junctions (Fig. 4C). One signaling pathway that may co-opt srGAP1 junctional recruitment is the hepatocyte growth factor (HGF) pathway. The HGF receptor, Met, is a receptor tyrosine kinase associated with partial epithelial–mesenchymal transition and loss of adherens junctions in cultured cells (Mangold et al., 2011). In vivo, Met is required for normal organogenesis in the liver and kidney, and high levels of HGF/Met are associated with poor cancer prognosis and metastasis (Trusolino et al., 2010). Although it is not likely that this is the only means by which HGF leads to reduced cell–cell adhesion, HGF-dependent reduction of RhoA activity is srGAP1-dependent in this system (Liang et al., 2017) (Fig. 4C). An earlier report connected HGF signaling to Slit2–Robo1 signaling via Rac1 in human cell lines; in this system, loss of Slit–Robo signaling led to increased Rac1 activation and exacerbated the effects of HGF (Stella et al., 2009). In the future, it will be interesting to see how various Slit–Robo signals and srGAPs combine to regulate HGF-dependent effects.
In addition to roles in cell–cell adhesion, srGAP1 may also play a role in regulating adhesion and migration of cells in fibrillar collagen matrices (Kutys and Yamada, 2014). A screen for guanine nucleotide exchange factors (GEFs) that regulate fibroblast migration on collagen identified βPix (also known as ARHGEF7), a dual-specificity GEF that regulates Cdc42 and Rac1 (Zhou et al., 2016). Knockdown of βPix leads to loss of polarized Cdc42 activity and blocks cell migration in fibrillar collagen microenvironments. Significantly, knockdown of Cdc42 or βPix, but not Rac1, leads to increased intracellular RhoA activity in fibroblasts migrating on fibrillar collagen. Surprisingly, co-immunoprecipitation and mass spectrometry analysis of βPix knockdown and βPix-expressing cells identified srGAP1 as a βPix-binding partner. Knockdown of srGAP1 phenocopies βPix or Cdc42 knockdown in 3D collagen matrices, suggesting that in addition to any direct roles in regulating Cdc42 in migration in collagen matrices, βPix may also mediate compensatory downregulation of RhoA by recruiting srGAP1 to sites of attachment to collagen.
These basic cell biological analyses of non-neuronal function of srGAPs agree well with a wealth of clinical studies, which have implicated srGAPs in non-neuronal disease (Box 2).
Box 2. srGAPs in non-neuronal disease.
Clinical roles for srGAPs outside of the nervous tissue have also been described. A gain-of-function mutation in srGAP1 and a mutation in Slit2 have been implicated in congenital anomalies of the kidney and urinary tract (CAKUT) (Hwang et al., 2015), consistent with disruption of the nephrogenic field in Robo2-knockout mice (Wainwright et al., 2015). srGAP1 and Arhgap4 are also candidate genes for regulating inflammatory responses. srGAP1 offers protection from neuroinflammation after brain injury in rats (Sherchan et al., 2016), and differential srGAP1 expression in leukocytes may regulate their differing chemotactic responses (Ye et al., 2010). Like the Slit–Robo system, which has been implicated in gastric (Huang et al., 2015) and pancreatic cancer (Bailey et al., 2016; Biankin et al., 2012), srGAPs have also been connected to cancer. An oncogenic fusion between srGAP3 and RAF1 is causative in some cases of pilocytic astrocytoma (Forshew et al., 2009; Jones et al., 2009), and mutations in a putative enhancer region of srGAP2 have been associated with increased breast cancer risk (Jiang et al., 2011). Some srGAP mutations in cancers lead to their inability to inactivate Rho GTPases, for example, mutations that inhibit the ability of srGAP1 to inactivate Cdc42 are associated with increased susceptibility to familial non-medullary thyroid carcinoma (He et al., 2013). srGAP3 expression is decreased in the majority of breast cancer cell lines, and loss of srGAP3 leads to both anchorage-independent growth and increased cell invasion and/or migration; these changes are GAP dependent, suggesting that srGAP3 acts as a tumor suppressor through its ability to inactivate Rac (Lahoz and Hall, 2013). Similar phenotypes are observed upon srGAP2 loss in osteosarcomas (Marko et al., 2016). Separating correlation from causation in the numerous clinical studies that have been reported will be challenging, but these studies clearly implicate srGAPs in a wide range of human disease.
Conclusions and perspectives
srGAPs play diverse roles in fundamental cell migratory and adhesive events, as well as during cancer and neural development. Continuing to dissect the functions of srGAPs within and outside of the nervous system will likely help to uncover potential treatments for a variety of disorders and diseases. Although members of the srGAP family have high sequence similarity and similar basic functions in membrane bending, actin regulation during cell migration, and neurodevelopment, their C-termini diverge considerably. A thorough investigation of C-terminal-binding partners for each of the srGAPs, as has been completed for srGAP3, would help to clarify the roles of srGAPs in various cell biological events. In particular, srGAP1 has emerged as an understudied protein, highlighting the need for future research efforts aimed at elucidating its roles in neuronal and non-neuronal tissues.
Given the identification of srGAPs as modulators of cadherin-dependent cell–cell adhesion in the C. elegans embryo (Zaidel-Bar et al., 2010) and in vertebrate tissue culture (Liang et al., 2017), further studies of the role of srGAPs in cell–cell adhesion events is also an exciting area for future investigation. While intriguing, the in vivo relevance of the proposed ‘protrusive activity buffer’ activity of srGAPs (Fritz et al., 2015) remains to be determined; one possible role for this function may be to modulate cell–cell adhesion. Another key unanswered question is the role of the C-terminus of srGAPs, known to be the nexus for numerous other protein–protein interactions, in cell–cell adhesion. Future studies aimed at elucidating its role may prove highly informative.
Finally, we are only beginning to understand, at a mechanistic level, how the misregulation of srGAP expression contributes to a wide variety of cancers. Understanding in more detail the normal expression patterns of srGAPs and elucidating how abnormalities in expression correlate with specific cancers may provide insights into both prognoses and potential best treatment paths for patients whose cancers are characterized by srGAP misexpression. Clearly, ‘minding’ the srGAPs will involve continued efforts not only to understand their roles in neurons and brains, but well beyond.
Acknowledgements
We thank A. Yap for sharing data prior to publication.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work of our laboratory was supported by the National Institutes of Health (R01 GM058038 to J.H.). B.L. was supported in part by a Genetics Training Grant from the NIH (T32 GM07133), and by a Michael J. Guyer fellowship through the University of Wisconsin-Madison Department of Integrative Biology. Deposited in PMC for release after 12 months.
References
- Abercrombie M. and Heaysman J. E. M. (1953). Observations on the social behaviour of cells in tissue culture. I. Speed of movement of chick heart fibroblasts in relation to their mutual contacts. Exp. Cell Res. 5, 111-131. 10.1016/0014-4827(53)90098-6 [DOI] [PubMed] [Google Scholar]
- Abercrombie M. and Heaysman J. E. M. (1954). Observations on the social behaviour of cells in tissue culture. II. Monolayering of fibroblasts. Exp. Cell Res. 6, 293-306. 10.1016/0014-4827(54)90176-7 [DOI] [PubMed] [Google Scholar]
- Almecija S., Wallace I. J., Judex S., Alba D. M. and Moya-Sola S. (2015). Human evolution. Comment on “Human-like hand use in Australopithecus africanus”. Science 348, 1101 10.1126/science.aaa8414 [DOI] [PubMed] [Google Scholar]
- Bacon C., Endris V. and Rappold G. (2009). Dynamic expression of the Slit-Robo GTPase activating protein genes during development of the murine nervous system. J. Comp. Neurol. 513, 224-236. 10.1002/cne.21955 [DOI] [PubMed] [Google Scholar]
- Bailey P., Chang D. K., Nones K., Johns A. L., Patch A.-M., Gingras M.-C., Miller D. K., Christ A. N., Bruxner T. J. C., Quinn M. C. et al. (2016). Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47-52. 10.1038/nature16965 [DOI] [PubMed] [Google Scholar]
- Bertram J., Koschützke L., Pfannmöller J. P., Esche J., van Diepen L., Kuss A. W., Hartmann B., Bartsch D., Lotze M. and von Bohlen Und Halbach O. (2016). Morphological and behavioral characterization of adult mice deficient for SrGAP3. Cell Tissue Res. 366, 1-11. 10.1007/s00441-016-2413-y [DOI] [PubMed] [Google Scholar]
- Biankin A. V., Waddell N., Kassahn K. S., Gingras M. C., Muthuswamy L. B., Johns A. L., Miller D. K., Wilson P. J., Patch A. M., Wu J. et al. (2012). Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399-405. 10.1038/nature11547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittel D. C., Kibiryeva N., Dasouki M., Knoll J. H. M. and Butler M. G. (2006). A 9-year-old male with a duplication of chromosome 3p25.3p26.2: clinical report and gene expression analysis. Am. J. Med. Genet. A 140, 573-579. 10.1002/ajmg.a.31132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga V. (2016). Spatial integration of E-cadherin adhesion, signalling and the epithelial cytoskeleton. Curr. Opin. Cell Biol. 42, 138-145. 10.1016/j.ceb.2016.07.006 [DOI] [PubMed] [Google Scholar]
- Carlson B. R., Lloyd K. E., Kruszewski A., Kim I.-H., Rodriguiz R. M., Heindel C., Faytell M., Dudek S. M., Wetsel W. C. and Soderling S. H. (2011). WRP/srGAP3 facilitates the initiation of spine development by an inverse F-BAR domain, and its loss impairs long-term memory. J. Neurosci. 31, 2447-2460. 10.1523/JNEUROSCI.4433-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Adler C. E., Krause M., Clark S. G., Gertler F. B., Tessier-Lavigne M. and Bargmann C. I. (2006). MIG-10/lamellipodin and AGE-1/PI3K promote axon guidance and outgrowth in response to slit and netrin. Curr. Biol. 16, 854-862. 10.1016/j.cub.2006.03.083 [DOI] [PubMed] [Google Scholar]
- Charrier C., Joshi K., Coutinho-Budd J., Kim J.-E., Lambert N., de Marchena J., Jin W.-L., Vanderhaeghen P., Ghosh A., Sassa T. et al. (2012). Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell 149, 923-935. 10.1016/j.cell.2012.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Mi Y.-J., Ma Y., Fu H.-L. and Jin W.-L. (2011). The mental retardation associated protein, srGAP3 negatively regulates VPA-induced neuronal differentiation of Neuro2A cells. Cell. Mol. Neurobiol. 31, 675-686. 10.1007/s10571-011-9664-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-P., Su Y.-N., Chen C.-Y., Su J.-W., Chern S.-R., Town D.-D. and Wang W. (2012a). Pure partial monosomy 3p (3p25.3 -> pter): prenatal diagnosis and array comparative genomic hybridization characterization. Taiwan. J. Obstet. Gynecol. 51, 435-439. 10.1016/j.tjog.2012.07.022 [DOI] [PubMed] [Google Scholar]
- Chen Z.-B., Zhang H.-Y., Zhao J.-H., Zhao W., Zhao D., Zheng L.-F., Zhang X.-F., Liao X.-P. and Yi X.-N. (2012b). Slit-Robo GTPase-activating proteins are differentially expressed in murine dorsal root ganglia: modulation by peripheral nerve injury. Anat. Rec. (Hoboken) 295, 652-660. 10.1002/ar.22419 [DOI] [PubMed] [Google Scholar]
- Chen Z., Chang K., Capraro B. R., Zhu C., Hsu C.-J. and Baumgart T. (2014). Intradimer/Intermolecular interactions suggest autoinhibition mechanism in endophilin A1. J. Am. Chem. Soc. 136, 4557-4564. 10.1021/ja411607b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christerson L. B., Gallagher E., Vanderbilt C. A., Whitehurst A. W., Wells C., Kazempour R., Sternweis P. C. and Cobb M. H. (2002). p115 Rho GTPase activating protein interacts with MEKK1. J. Cell. Physiol. 192, 200-208. 10.1002/jcp.10125 [DOI] [PubMed] [Google Scholar]
- Coutinho-Budd J., Ghukasyan V., Zylka M. J. and Polleux F. (2012). The F-BAR domains from srGAP1, srGAP2 and srGAP3 regulate membrane deformation differently. J. Cell Sci. 125, 3390-3401. 10.1242/jcs.098962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y.-K., Ma Y., Chen K., Mi Y.-J., Fu H.-L., Cui D.-X. and Jin W.-L. (2014). A link between the nuclear-localized srGAP3 and the SWI/SNF chromatin remodeler Brg1. Mol. Cell. Neurosci. 60, 10-25. 10.1016/j.mcn.2014.02.005 [DOI] [PubMed] [Google Scholar]
- Dennis M. Y., Nuttle X., Sudmant P. H., Antonacci F., Graves T. A., Nefedov M., Rosenfeld J. A., Sajjadian S., Malig M., Kotkiewicz H. et al. (2012). Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 149, 912-922. 10.1016/j.cell.2012.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellery P. M., Ellis R. J. and Holder S. E. (2014). Interstitial 3p25 deletion in a patient with features of 3p deletion syndrome: further evidence for the role of SRGAP3 in mental retardation. Clin. Dysmorphol. 23, 29-31. 10.1097/MCD.0000000000000017 [DOI] [PubMed] [Google Scholar]
- Endris V., Wogatzky B., Leimer U., Bartsch D., Zatyka M., Latif F., Maher E. R., Tariverdian G., Kirsch S., Karch D. et al. (2002). The novel Rho-GTPase activating gene MEGAP/ srGAP3 has a putative role in severe mental retardation. Proc. Natl. Acad. Sci. USA 99, 11754-11759. 10.1073/pnas.162241099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endris V., Haussmann L., Buss E., Bacon C., Bartsch D. and Rappold G. (2011). SrGAP3 interacts with lamellipodin at the cell membrane and regulates Rac-dependent cellular protrusions. J. Cell Sci. 124, 3941-3955. 10.1242/jcs.077081 [DOI] [PubMed] [Google Scholar]
- Foletta V. C., Brown F. D. and Young W. S. III (2002). Cloning of rat ARHGAP4/C1, a RhoGAP family member expressed in the nervous system that colocalizes with the Golgi complex and microtubules. Brain Res. Mol. Brain Res. 107, 65-79. 10.1016/S0169-328X(02)00448-5 [DOI] [PubMed] [Google Scholar]
- Forshew T., Tatevossian R. G., Lawson A. R. J., Ma J., Neale G., Ogunkolade B. W., Jones T. A., Aarum J., Dalton J., Bailey S. et al. (2009). Activation of the ERK/MAPK pathway: a signature genetic defect in posterior fossa pilocytic astrocytomas. J. Pathol. 218, 172-181. 10.1002/path.2558 [DOI] [PubMed] [Google Scholar]
- Fossati M., Pizzarelli R., Schmidt E. R., Kupferman J. V., Stroebel D., Polleux F. and Charrier C. (2016). SRGAP2 and its human-specific paralog co-regulate the development of excitatory and inhibitory synapses. Neuron 91, 356-369. 10.1016/j.neuron.2016.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz R. D., Menshykau D., Martin K., Reimann A., Pontelli V. and Pertz O. (2015). SrGAP2-dependent integration of membrane geometry and slit-robo-repulsive cues regulates fibroblast contact inhibition of locomotion. Dev. Cell 35, 78-92. 10.1016/j.devcel.2015.09.002 [DOI] [PubMed] [Google Scholar]
- Frost A., Perera R., Roux A., Spasov K., Destaing O., Egelman E. H., De Camilli P. and Unger V. M. (2008). Structural basis of membrane invagination by F-BAR domains. Cell 132, 807-817. 10.1016/j.cell.2007.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Ma Y., Fu H.-L., Luo Q., Wang Z., Xiao Y.-H., Yang H., Cui D.-X. and Jin W.-L. (2016). Non-catalytic roles for TET1 protein negatively regulating neuronal differentiation through srGAP3 in neuroblastoma cells. Protein Cell 7, 351-361. 10.1007/s13238-016-0267-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier S., Coutinho-Budd J., Sassa T., Gresset A., Jordan N. V., Chen K., Jin W.-L., Frost A. and Polleux F. (2009). The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 138, 990-1004. 10.1016/j.cell.2009.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guez-Haddad J., Sporny M., Sasson Y., Gevorkyan-Airapetov L., Lahav-Mankovski N., Margulies D., Radzimanowski J. and Opatowsky Y. (2015). The neuronal migration factor srGAP2 achieves specificity in ligand binding through a two-component molecular mechanism. Structure 23, 1989-2000. 10.1016/j.str.2015.08.009 [DOI] [PubMed] [Google Scholar]
- Gunnarsson C. and Foyn Bruun C. (2010). Molecular characterization and clinical features of a patient with an interstitial deletion of 3p25.3-p26.1. Am. J. Med. Genet. A 152A, 3110-3114. 10.1002/ajmg.a.33353 [DOI] [PubMed] [Google Scholar]
- Guo S. and Bao S. (2010). srGAP2 arginine methylation regulates cell migration and cell spreading through promoting dimerization. J. Biol. Chem. 285, 35133-35141. 10.1074/jbc.M110.153429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S. D. and Mullins R. D. (2015). Lamellipodin promotes actin assembly by clustering Ena/VASP proteins and tethering them to actin filaments. Elife 4, e06585 10.7554/eLife.06585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J. C., Yu T. W., Fujisawa K., Culotti J. G., Gengyo-Ando K., Mitani S., Moulder G., Barstead R., Tessier-Lavigne M. and Bargmann C. I. (2001). C. elegans slit acts in midline, dorsal-ventral, and anterior-posterior guidance via the SAX-3/Robo receptor. Neuron 32, 25-38. 10.1016/S0896-6273(01)00448-2 [DOI] [PubMed] [Google Scholar]
- He H., Bronisz A., Liyanarachchi S., Nagy R., Li W., Huang Y., Akagi K., Saji M., Kula D., Wojcicka A. et al. (2013). SRGAP1 is a candidate gene for papillary thyroid carcinoma susceptibility. J. Clin. Endocrinol. Metab. 98, E973-E980. 10.1210/jc.2012-3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho N. T., Kim P. S., Kutzner A. and Heese K. (2017). Cognitive functions: human vs. animal - 4:1 advantage -|FAM72-SRGAP2-|. J. Mol. Neurosci. 61, 603-606. 10.1007/s12031-017-0901-5 [DOI] [PubMed] [Google Scholar]
- Huang W., Ciochon R., Gu Y., Larick R., Qiren F., Schwarcz H., Yonge C., de Vos J. and Rink W. (1995). Early Homo and associated artefacts from Asia. Nature 378, 275-278. 10.1038/378292a0 [DOI] [PubMed] [Google Scholar]
- Huang T., Kang W., Cheng A. S. L., Yu J. and To K. F. (2015). The emerging role of Slit-Robo pathway in gastric and other gastro intestinal cancers. BMC Cancer 15, 950 10.1186/s12885-015-1984-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D.-Y., Kohl S., Fan X., Vivante A., Chan S., Dworschak G. C., Schulz J., van Eerde A. M., Hilger A. C., Gee H. Y. et al. (2015). Mutations of the SLIT2-ROBO2 pathway genes SLIT2 and SRGAP1 confer risk for congenital anomalies of the kidney and urinary tract. Hum. Genet. 134, 905-916. 10.1007/s00439-015-1570-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarsch I. K., Daste F. and Gallop J. L. (2016). Membrane curvature in cell biology: an integration of molecular mechanisms. J. Cell Biol. 214, 375-387. 10.1083/jcb.201604003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Shen H., Liu X., Dai J., Jin G., Qin Z., Chen J., Wang S., Wang X., Hu Z. et al. (2011). Genetic variants at 1p11.2 and breast cancer risk: a two-stage study in Chinese women. PLoS ONE 6, e21563 10.1371/journal.pone.0021563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Q., Wang L., Zhang Z., Wang Y., Yan H., Ma W., Jin W., Lu H. and Liu Y. (2016). Dynamic expression of srGAP2 in cell nuclei and cytoplasm during the differentiation of rat neural stem cells in vitro. Mol. Med. Rep. 14, 4599-4605. 10.3892/mmr.2016.5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T. W., Kocialkowski S., Liu L., Pearson D. M., Ichimura K. and Collins V. P. (2009). Oncogenic RAF1 rearrangement and a novel BRAF mutation as alternatives to KIAA1549:BRAF fusion in activating the MAPK pathway in pilocytic astrocytoma. Oncogene 28, 2119-2123. 10.1038/onc.2009.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley C. F., Messelaar E. M., Eskin T. L., Wang S., Song K., Vishnia K., Becalska A. N., Shupliakov O., Hagan M. F., Danino D. et al. (2015). Membrane charge directs the outcome of F-BAR domain lipid binding and autoregulation. Cell Rep. 13, 2597-2609. 10.1016/j.celrep.2015.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessels M. M. and Qualmann B. (2015). Different functional modes of BAR domain proteins in formation and plasticity of mammalian postsynapses. J. Cell Sci. 128, 3177-3185. 10.1242/jcs.174193 [DOI] [PubMed] [Google Scholar]
- Kim I. H., Carlson B. R., Heindel C. C., Kim H. and Soderling S. H. (2012). Disruption of wave-associated Rac GTPase-activating protein (Wrp) leads to abnormal adult neural progenitor migration associated with hydrocephalus. J. Biol. Chem. 287, 39263-39274. 10.1074/jbc.M112.398834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschützke L., Bertram J., Hartmann B., Bartsch D., Lotze M. and von Bohlen und Halbach O. (2015). SrGAP3 knockout mice display enlarged lateral ventricles and specific cilia disturbances of ependymal cells in the third ventricle. Cell Tissue Res. 361, 645-650. 10.1007/s00441-015-2224-6 [DOI] [PubMed] [Google Scholar]
- Krause M., Leslie J. D., Stewart M., Lafuente E. M., Valderrama F., Jagannathan R., Strasser G. A., Rubinson D. A., Liu H., Way M. et al. (2004). Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev. Cell 7, 571-583. 10.1016/j.devcel.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Kutys M. L. and Yamada K. M (2014). An extracellular-matrix-specific GEF-GAP interaction regulates Rho GTPase crosstalk for 3D collagen migration. Nat. Cell Biol. 16, 909-917. 10.1038/ncb3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahoz A. and Hall A. (2013). A tumor suppressor role for srGAP3 in mammary epithelial cells. Oncogene 32, 4854-4860. 10.1038/onc.2012.489 [DOI] [PubMed] [Google Scholar]
- Lei W., Omotade O. F., Myers K. R. and Zheng J. Q. (2016). Actin cytoskeleton in dendritic spine development and plasticity. Curr. Opin. Neurobiol. 39, 86-92. 10.1016/j.conb.2016.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Budnar S., Gupta S., Verma S., Han S. P., Daly R. J., Parton R. G., Hamilton N. A., Gomez G. A. and Yap A. S. (2017). Tyrosine dephosphorylated cortactin downregulates contractility at the epithelial zonula adherens through SRGAP1. Nat. Commun. 8, 790. 10.1038/s41467-017-00797-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Jiao Q., Wang Y., Yang Z., Feng M., Wang L., Chen X., Jin W. and Liu Y. (2013). The mental retardation-associated protein srGAP3 regulates survival, proliferation, and differentiation of rat embryonic neural stem/progenitor cells. Stem Cells Dev. 22, 1709-1716. 10.1089/scd.2012.0455 [DOI] [PubMed] [Google Scholar]
- Ma Y., Mi Y.-J., Dai Y.-K., Fu H.-L., Cui D.-X. and Jin W.-L. (2013). The inverse F-BAR domain protein srGAP2 acts through srGAP3 to modulate neuronal differentiation and neurite outgrowth of mouse neuroblastoma cells. PLoS ONE 8, e57865 10.1371/journal.pone.0057865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold S., Wu S. K., Norwood S. J., Collins B. M., Hamilton N. A., Thorn P. and Yap A. S. (2011). Hepatocyte growth factor acutely perturbs actin filament anchorage at the epithelial zonula adherens. Curr. Biol. 21, 503-507. 10.1016/j.cub.2011.02.018 [DOI] [PubMed] [Google Scholar]
- Marko T. A., Shamsan G. A., Edwards E. N., Hazelton P. E., Rathe S. K., Cornax I., Overn P. R., Varshney J., Diessner B. J., Moriarity B. S. et al. (2016). Slit-Robo GTPase-activating protein 2 as a metastasis suppressor in osteosarcoma. Sci. Rep. 6, 39059 10.1038/srep39059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason F. M., Heimsath E. G., Higgs H. N. and Soderling S. H. (2011). Bi-modal regulation of a formin by srGAP2. J. Biol. Chem. 286, 6577-6586. 10.1074/jbc.M110.190397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar P., Britz O., Johannes C., Nieto M., Ma L., Rebeyka A., Klenin N., Polleux F., Guillemot F. and Schuurmans C. (2004). A screen for downstream effectors of Neurogenin2 in the embryonic neocortex. Dev. Biol. 273, 373-389. 10.1016/j.ydbio.2004.06.013 [DOI] [PubMed] [Google Scholar]
- McMahon H. T. and Boucrot E. (2015). Membrane curvature at a glance. J. Cell Sci. 128, 1065-1070. 10.1242/jcs.114454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor H. (2010). The role of formins in filopodia formation. Biochim. Biophys. Acta 1803, 191-200. 10.1016/j.bbamcr.2008.12.018 [DOI] [PubMed] [Google Scholar]
- Mim C. and Unger V. M. (2012). Membrane curvature and its generation by BAR proteins. Trends Biochem. Sci. 37, 526-533. 10.1016/j.tibs.2012.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera-de Benito D., Garcia-Perez M. A., Martinez-Granero M. A. and Izquierdo-Lopez L. (2014). A patient with a duplication of chromosome 3p (p24.1p26.2): a comparison with other partial 3p trisomies. Am. J. Med. Genet. A 164A, 548-550. 10.1002/ajmg.a.36164 [DOI] [PubMed] [Google Scholar]
- Neukomm L. J., Frei A. P., Cabello J., Kinchen J. M., Zaidel-Bar R., Ma Z., Haney L. B., Hardin J., Ravichandran K. S., Moreno S. et al. (2011). Loss of the RhoGAP SRGP-1 promotes the clearance of dead and injured cells in Caenorhabditis elegans. Nat. Cell Biol. 13, 79-86. 10.1038/ncb2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukomm L. J., Zeng S., Frei A. P., Huegli P. A. and Hengartner M. O. (2014). Small GTPase CDC-42 promotes apoptotic cell corpse clearance in response to PAT-2 and CED-1 in C. elegans. Cell Death Differ. 21, 845-853. 10.1038/cdd.2014.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttle X., Huddleston J., O'Roak B. J., Antonacci F., Fichera M., Romano C., Shendure J. and Eichler E. E. (2013). Rapid and accurate large-scale genotyping of duplicated genes and discovery of interlocus gene conversions. Nat. Methods 10, 903-909. 10.1038/nmeth.2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltekova I. T., Macdonald A. and Armour C. M. (2012). Microdeletion on 3p25 in a patient with features of 3p deletion syndrome. Am. J. Med. Genet. A 158A, 2583-2586. 10.1002/ajmg.a.35559 [DOI] [PubMed] [Google Scholar]
- Quinn C. C., Pfeil D. S. and Wadsworth W. G. (2008). CED-10/Rac1 mediates axon guidance by regulating the asymmetric distribution of MIG-10/lamellipodin. Curr. Biol. 18, 808-813. 10.1016/j.cub.2008.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers G. J. A. (2002). Rho proteins, mental retardation and the cellular basis of cognition. Trends Neurosci. 25, 191-199. 10.1016/S0166-2236(00)02118-4 [DOI] [PubMed] [Google Scholar]
- Rao Y., Ma Q., Vahedi-Faridi A., Sundborger A., Pechstein A., Puchkov D., Luo L., Shupliakov O., Saenger W. and Haucke V. (2010). Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation. Proc. Natl. Acad. Sci. USA 107, 8213-8218. 10.1073/pnas.1003478107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratheesh A., Priya R. and Yap A. S. (2013). Coordinating Rho and Rac: the regulation of Rho GTPase signaling and cadherin junctions. Prog. Mol. Biol. Transl. Sci. 116, 49-68. 10.1016/B978-0-12-394311-8.00003-0 [DOI] [PubMed] [Google Scholar]
- Riess A., Grasshoff U., Schäferhoff K., Bonin M., Riess O., Horber V. and Tzschach A. (2012). Interstitial 3p25.3-p26.1 deletion in a patient with intellectual disability. Am. J. Med. Genet. A 158A, 2587-2590. 10.1002/ajmg.a.35562 [DOI] [PubMed] [Google Scholar]
- Rincic M., Rados M., Krsnik Z., Gotovac K., Borovecki F., Liehr T. and Brecevic L. (2016). Complex intrachromosomal rearrangement in 1q leading to 1q32.2 microdeletion: a potential role of SRGAP2 in the gyrification of cerebral cortex. Mol. Cytogenet. 9, 19 10.1186/s13039-016-0221-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochefort N. L. and Konnerth A. (2012). Dendritic spines: from structure to in vivo function. EMBO Rep. 13, 699-708. 10.1038/embor.2012.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitsu H., Osaka H., Sugiyama S., Kurosawa K., Mizuguchi T., Nishiyama K., Nishimura A., Tsurusaki Y., Doi H., Miyake N. et al. (2012). Early infantile epileptic encephalopathy associated with the disrupted gene encoding Slit-Robo Rho GTPase activating protein 2 (SRGAP2). Am. J. Med. Genet. A 158A, 199-205. 10.1002/ajmg.a.34363 [DOI] [PubMed] [Google Scholar]
- Schwyzer U., Binkert F., Caflisch U., Baumgartner B. and Schinzel A. (1987). Terminal deletion of the short arm of chromosome 3, del(3pter-p25): a recognizable syndrome. Helv. Paediatr. Acta 42, 309-315. [PubMed] [Google Scholar]
- Sherchan P., Huang L., Wang Y., Akyol O., Tang J. and Zhang J. H. (2016). Recombinant Slit2 attenuates neuroinflammation after surgical brain injury by inhibiting peripheral immune cell infiltration via Robo1-srGAP1 pathway in a rat model. Neurobiol. Dis. 85, 164-173. 10.1016/j.nbd.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuib S., McMullan D., Rattenberry E., Barber R. M., Rahman F., Zatyka M., Chapman C., Macdonald F., Latif F., Davison V. et al. (2009). Microarray based analysis of 3p25-p26 deletions (3p- syndrome). Am. J. Med. Genet. A 149A, 2099-2105. 10.1002/ajmg.a.32824 [DOI] [PubMed] [Google Scholar]
- Skinner M. M., Stephens N. B., Tsegai Z. J., Foote A. C., Nguyen N. H., Gross T., Pahr D. H., Hublin J. J. and Kivell T. L. (2015a). Human evolution. Human-like hand use in Australopithecus africanus. Science 347, 395-399. 10.1126/science.1261735 [DOI] [PubMed] [Google Scholar]
- Skinner M. M., Stephens N. B., Tsegai Z. J., Foote A. C., Nguyen N. H., Gross T., Pahr D. H., Hublin J. J. and Kivell T. L. (2015b). Human evolution. Response to Comment on “Human-like hand use in Australopithecus africanus”. Science 348, 1101 10.1126/science.aaa8931 [DOI] [PubMed] [Google Scholar]
- Soderling S. H., Binns K. L., Wayman G. A., Davee S. M., Ong S. H., Pawson T. and Scott J. D. (2002). The WRP component of the WAVE-1 complex attenuates Rac-mediated signalling. Nat. Cell Biol. 4, 970-975. 10.1038/ncb886 [DOI] [PubMed] [Google Scholar]
- Soderling S. H., Langeberg L. K., Soderling J. A., Davee S. M., Simerly R., Raber J. and Scott J. D. (2003). Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc. Natl. Acad. Sci. USA 100, 1723-1728. 10.1073/pnas.0438033100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling S. H., Guire E. S., Kaech S., White J., Zhang F., Schutz K., Langeberg L. K., Banker G., Raber J. and Scott J. D. (2007). A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J. Neurosci. 27, 355-365. 10.1523/JNEUROSCI.3209-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporny M., Guez-Haddad J., Waterman D. G., Isupov M. N. and Opatowsky Y. (2016). Molecular symmetry-constrained systematic search approach to structure solution of the coiled-coil SRGAP2 F-BARx domain. Acta Crystallogr. D Struct. Biol. 72, 1241-1253. 10.1107/S2059798316016697 [DOI] [PubMed] [Google Scholar]
- Sporny M., Guez-Haddad J., Kreusch A., Shakartzi S., Neznansky A., Cross A., Isupov M. N., Qualmann B., Kessels M. M. and Opatowsky Y. (2017). Structural history of human SRGAP2 proteins. Mol. Biol. Evol. 34, 1463-1478. 10.1093/molbev/msx094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanishneva-Konovalova T. B., Kelley C. F., Eskin T. L., Messelaar E. M., Wasserman S. A., Sokolova O. S. and Rodal A. A. (2016). Coordinated autoinhibition of F-BAR domain membrane binding and WASp activation by nervous wreck. Proc. Natl. Acad. Sci. USA 113, E5552-E5561. 10.1073/pnas.1524412113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella M. C., Trusolino L. and Comoglio P. M. (2009). The Slit/Robo system suppresses hepatocyte growth factor-dependent invasion and morphogenesis. Mol. Biol. Cell 20, 642-657. 10.1091/mbc.E08-03-0321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. C., McPherron S. P., Bobe R., Reed D., Barr W. A., Wynn J. G., Marean C. W., Geraads D. and Alemseged Z. (2015). Taphonomy of fossils from the hominin-bearing deposits at Dikika, Ethiopia. J. Hum. Evol. 86, 112-135. 10.1016/j.jhevol.2015.06.013 [DOI] [PubMed] [Google Scholar]
- Trusolino L., Bertotti A. and Comoglio P. M. (2010). MET signalling: principles and functions in development, organ regeneration and cancer. Nat. Rev. Mol. Cell Biol. 11, 834-848. 10.1038/nrm3012 [DOI] [PubMed] [Google Scholar]
- Vázquez F. X., Unger V. M. and Voth G. A. (2013). Autoinhibition of endophilin in solution via interdomain interactions. Biophys. J. 104, 396-403. 10.1016/j.bpj.2012.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verjaal M. and De Nef M. B. (1978). A patient with a partial deletion of the short arm of chromosome 3. Am. J. Dis. Child 132, 43-45. 10.1001/archpedi.1978.02120260045012 [DOI] [PubMed] [Google Scholar]
- Vogt D. L., Gray C. D., Young W. S., Orellana S. A. and Malouf A. T. (2007). ARHGAP4 is a novel RhoGAP that mediates inhibition of cell motility and axon outgrowth. Mol. Cell. Neurosci. 36, 332-342. 10.1016/j.mcn.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright E. N., Wilhelm D., Combes A. N., Little M. H. and Koopman P. (2015). ROBO2 restricts the nephrogenic field and regulates Wolffian duct-nephrogenic cord separation. Dev. Biol. 404, 88-102. 10.1016/j.ydbio.2015.05.023 [DOI] [PubMed] [Google Scholar]
- Waltereit R., Kautt S. and Bartsch D. (2008). Expression of MEGAP mRNA during embryonic development. Gene Expr. Patterns 8, 307-310. 10.1016/j.gep.2008.01.006 [DOI] [PubMed] [Google Scholar]
- Waltereit R., Leimer U., von Bohlen und Halbach O., Panke J., Holter S. M., Garrett L., Wittig K., Schneider M., Schmitt C., Calzada-Wack J. et al. (2012). Srgap3(−/−) mice present a neurodevelopmental disorder with schizophrenia-related intermediate phenotypes. FASEB J. 26, 4418-4428. 10.1096/fj.11-202317 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhang Y., Zhang Z., Jin W.-L. and Wu G. (2014). Purification, crystallization and preliminary X-ray analysis of the inverse F-BAR domain of the human srGAP2 protein. Acta Crystallogr. F Struct. Biol. Commun. 70, 123-126. 10.1107/S2053230X13033712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K., Ren X.-R., Huang Y.-Z., Xie Y., Liu G., Saito H., Tang H., Wen L., Brady-Kalnay S. M., Mei L. et al. (2001). Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit-Robo pathway. Cell 107, 209-221. 10.1016/S0092-8674(01)00530-X [DOI] [PubMed] [Google Scholar]
- Wu T. and Baumgart T. (2014). BIN1 membrane curvature sensing and generation show autoinhibition regulated by downstream ligands and PI(4,5)P2. Biochemistry 53, 7297-7309. 10.1021/bi501082r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuertenberger S. and Groemping Y. (2015). A single PXXP motif in the C-terminal region of srGAP3 mediates binding to multiple SH3 domains. FEBS Lett. 589, 1156-1163. 10.1016/j.febslet.2015.03.014 [DOI] [PubMed] [Google Scholar]
- Yamada S. and Nelson W. J. (2007). Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell-cell adhesion. J. Cell Biol. 178, 517-527. 10.1083/jcb.200701058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D., Itoh T., Miki H. and Takenawa T. (2013). srGAP1 regulates lamellipodial dynamics and cell migratory behavior by modulating Rac1 activity. Mol. Biol. Cell 24, 3393-3405. 10.1091/mbc.E13-04-0178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Marcello M., Endris V., Saffrich R., Fischer R., Trendelenburg M. F., Sprengel R. and Rappold G. (2006). MEGAP impedes cell migration via regulating actin and microtubule dynamics and focal complex formation. Exp. Cell Res. 312, 2379-2393. 10.1016/j.yexcr.2006.04.001 [DOI] [PubMed] [Google Scholar]
- Ye B.-Q., Geng Z. H., Ma L. and Geng J.-G. (2010). Slit2 regulates attractive eosinophil and repulsive neutrophil chemotaxis through differential srGAP1 expression during lung inflammation. J. Immunol. 185, 6294-6305. 10.4049/jimmunol.1001648 [DOI] [PubMed] [Google Scholar]
- Ypsilanti A. R., Zagar Y. and Chedotal A. (2010). Moving away from the midline: new developments for Slit and Robo. Development 137, 1939-1952. 10.1242/dev.044511 [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R., Joyce M. J., Lynch A. M., Witte K., Audhya A. and Hardin J. (2010). The F-BAR domain of SRGP-1 facilitates cell-cell adhesion during C. elegans morphogenesis. J. Cell Biol. 191, 761-769. 10.1083/jcb.201005082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen J. A., Yi B. A. and Bargmann C. I. (1998). The conserved immunoglobulin superfamily member SAX-3/Robo directs multiple aspects of axon guidance in C. elegans. Cell 92, 217-227. 10.1016/S0092-8674(00)80916-2 [DOI] [PubMed] [Google Scholar]
- Zhou W., Li X. and Premont R. T (2016). Expanding functions of GIT Arf GTPase-activating proteins, PIX Rho guanine nucleotide exchange factors and GIT-PIX complexes. J. Cell Sci. 129, 1963-1974. 10.1242/jcs.179465 [DOI] [PMC free article] [PubMed] [Google Scholar]