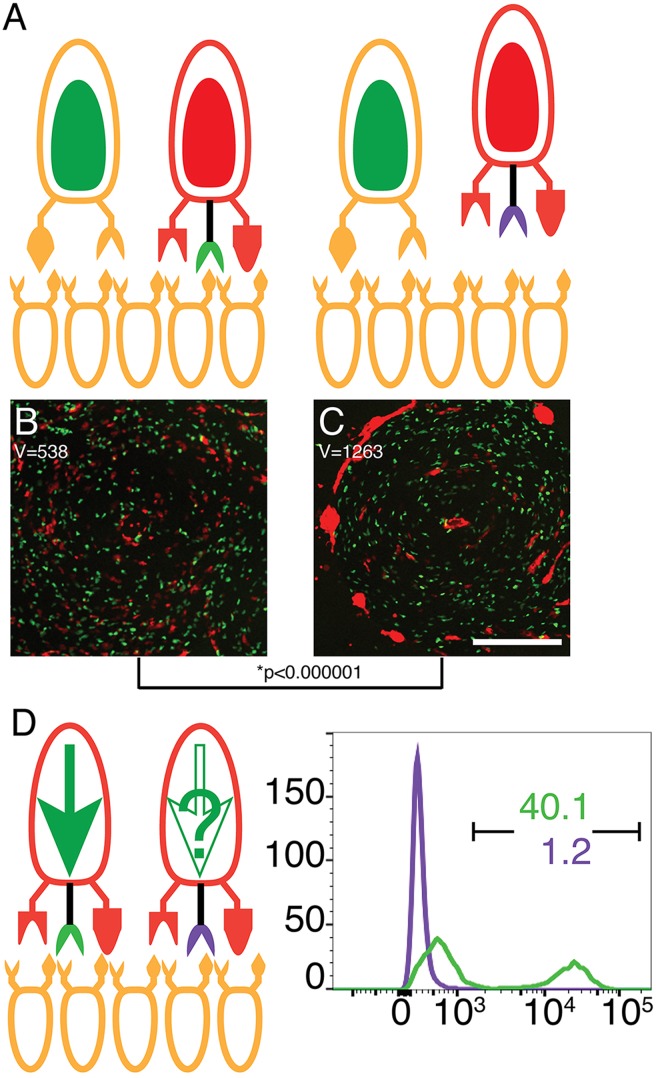
Fig. 3.
The cytoplasmic domain of TgrB1 is required for cooperative aggregation and differentiation. (A–C) Ovoids represent cells, protrusions represent TgrB1 and TgrC1 proteins, and colors represent allotypes: AX4 (tan) and tgrB1C1QS31 (red). Black protrusions represent extra alleles: tgrB1AX4-HA (green) and tgrB1AX4Δ828-902-HA (purple). Cooperative aggregation of mixes of 90% unlabeled AX4 cells with 5% GFP-labeled AX4 and 5% RFP-labeled tgrB1C1QS31tgrB1AX4-HA (B) or tgrB1C1QS31tgrB1AX4Δ828-902-HA (C). The spatial distribution variance (V) is shown inside each frame. A comparison between the variances is shown below the images with a P-value calculated from F-tests; *P<0.000001. Scale bar: 0.3 mm. (D) Cooperative differentiation of mixes of 0.2% cells carrying cotB-GFP with 99.8% unlabeled AX4 cells. Protrusions represent TgrB1 and TgrC1 and the colors represent alleles as above. Full green arrow inside the cells shows cotB-GFP expression; empty arrow indicates no expression. We evaluated GFP fluorescence levels by flow cytometry and plotted the results as histograms. The x-axis represents fluorescence intensify (arbitrary units) and the y-axis the number of events. The black bar indicates the GFP-positive populations and the numbers indicate the respective fractions (%) of GFP-positive cells.