Abstract
Many pigment cells acquire unique structural properties and gene expression profiles during animal development. The underlying differentiation pathways have been well characterized in cells formed during embryogenesis, such as the neural crest-derived melanocyte. However, much less is known about the developmental origins of pigment cells produced in adult organisms during tissue homeostasis and repair. Here we report a lineage analysis of ommochrome- and porphyrin-producing cells in the brown, freshwater planarian Schmidtea mediterranea. Using an RNA-sequencing approach, we identified two classes of markers expressed in sequential fashion when new pigment cells are generated during regeneration or in response to pigment cell ablation. We also report roles for FOXF-1 and ETS-1 transcription factors, as well as for an FGFR-like molecule, in the specification and maintenance of this cell type. Together, our results provide insights into mechanisms of adult pigment cell development in the strikingly colorful Platyhelminthes phylum.
KEY WORDS: Planarian, Pigment cell, Schmidtea mediterranea, Regeneration
Summary: Identification of the dendritic and punctate classes of marker and of foxF-1, ets-1 and fgfrL-1 as novel specification and maintenance regulators provides insight into mechanisms of pigment cell development and regeneration in adult Schmidtea mediterranea.
INTRODUCTION
The diverse colors of life derive from organic pigments that selectively absorb particular wavelengths of light. These molecules are often produced by specialized cells that acquire unique structural and functional properties during embryonic development. Most vertebrate pigment cells, for example, arise from the neural crest (NC), a multipotent population of cells that delaminate from the neuroepithelium and migrate throughout the embryo to adopt a wide variety of fates (Pavan and Raible, 2012). A subset of NC cells undergo dorsolateral migration away from the neural tube, colonize the overlying ectoderm, and differentiate to form melanocytes in birds and mammals or chromatophores in amphibians, reptiles and fish (Mills and Patterson, 2009). Melanocytes produce the dark, tyrosine-derived pigment melanin, and then transport it via dendrites into neighboring keratinocytes, where it protects DNA from ultraviolet radiation (Cichorek et al., 2013). Melanocytes also impart color to feathers and hair. Chromatophores synthesize a variety of pigments, including melanin (melanophores), carotenoids and/or pteridines (erythrophores and xanthophores) and purines (iridophores), but generally retain them intracellularly rather than distributing them to other cell types (Eom et al., 2015; Mills and Patterson, 2009). Many chromatophores also exhibit a dendritic morphology, facilitating regulated aggregation and dispersal of pigment granules to effect rapid color changes for purposes such as camouflage (Aspengren et al., 2009).
Some animals retain the capacity to generate new pigment cells beyond embryogenesis. In zebrafish, for instance, the alternating horizontal stripes of the skin are formed from black melanophores and yellow-orange xanthophores, with iridescent iridophores conferring a shiny appearance (Lister, 2002). Xanthophores are derived directly from embryonic NC cells, while the melanophore and iridophore lineages include larval or adult stem cells associated with the peripheral nervous system (Parichy and Spiewak, 2015). Larval melanophores can be reformed from otherwise quiescent stem cells following ablation (Yang and Johnson, 2006), and new melanophores also arise from undifferentiated precursors during fin regeneration in adults (Rawls and Johnson, 2000). Similarly, adult stem cells at the base of hair follicles drive melanocyte production during the hair regeneration cycle in mammals (Nishimura et al., 2002).
Despite the fact that melanocytes are by far the most widely studied pigment cells, numerous aspects of their biology remain unknown. For example, they have been found in the inner ear, heart and brain, in addition to the epidermis (Goldgeier et al., 1984; Mjaatvedt et al., 2005; Roberts and Linthicum, 2015), and their lineage was recently shown to encompass ventrally migrating NC cells better known for generating Schwann cells (Adameyko et al., 2009). Even the widely cited photoprotective function of melanin in the skin might be an oversimplification, given several recent studies indicating that it can act as a potent photosensitizer (Chiarelli-Neto et al., 2014; Noonan et al., 2012; Premi et al., 2015; Takeuchi et al., 2004). Although recent progress has been made in organisms such as zebrafish (Parichy and Spiewak, 2015), non-melanocyte pigment cells and the molecular mechanisms that regulate their development are even less well understood. We have much to learn, therefore, about the biology of organic pigments and the cells that produce them.
Here we conducted a lineage analysis of pigment cells in adult freshwater planarians (lophotrochozoan flatworms) (Newmark and Sanchez Alvarado, 2002; Reddien and Sanchez Alvarado, 2004). Like many other animals, planarians exhibit highly variable pigmentation between species (Hyman, 1951). Biochemical and genetic evidence indicate that the body pigment in Schmidtea mediterranea is composed of ommochromes and cyclic tetrapyrroles called porphyrins (Hase et al., 2006; Stubenhaus et al., 2016). Enzymes in the ommochrome and porphyrin biosynthesis pathways are expressed in a candidate pigment cell population that is highly dendritic in morphology, and RNAi knockdown of some of these genes led to white animals (Stubenhaus et al., 2016; Wang et al., 2016). Porphyrins react with oxygen to form free radicals when exposed to light. Thus, prolonged light exposure ablates S. mediterranea pigment cells, resulting in an otherwise healthy, white animal (Stubenhaus et al., 2016).
In the current study, we take a systematic approach to defining the dynamics and regulation of the S. mediterranea pigment cell lineage by performing whole-animal mRNA sequencing (RNAseq) at multiple time points during light-induced depigmentation and subsequent repigmentation. This analysis revealed ten pigment cell markers that can be divided into two general categories: ʻdendritic' markers exhibit a unique expression pattern revealing the highly arborized morphology of the pigment cells; whereas the more numerous ʻpunctate' markers exhibit more focused RNA localization that is likely to reflect confinement to the cell body. Both categories of markers are expressed in the same subepidermal space and exhibit some degree of overlap at steady state, suggesting that they are co-expressed in the same cell type. When animals were challenged to make pigment cells de novo during regeneration or repigmentation of depigmented animals, dendritic markers appeared first, suggesting that they are involved in pigment biosynthesis pathways activated early during pigment cell differentiation. Finally, using single-cell RNAseq (scRNAseq) datasets, we identified three novel regulators of pigment cells: foxF-1, ets-1, and an FGFR-like molecule fgfrL-1. Our work provides the first molecular insight into pigment cell lineage specification in planarians.
RESULTS
RNAseq analysis identifies two classes of pigment cell markers
In order to discover transcripts expressed in planarian pigment cells, we used RNAseq to compare gene expression in fully pigmented control animals, fully depigmented animals obtained by prolonged light exposure (Stubenhaus et al., 2016), and animals beginning to repigment after being returned to a dark environment (Fig. 1A; see Materials and Methods). In total, 436 transcripts were found to decrease by ≥2-fold with depigmentation (Table S1), 75 of which were cloned and screened by whole-mount in situ hybridization (WISH) (Fig. S1) for patterns consistent with pigment cell-specific or enriched expression (Stubenhaus et al., 2016; Wang et al., 2016). From this screen, two classes of pigment cell markers were identified.
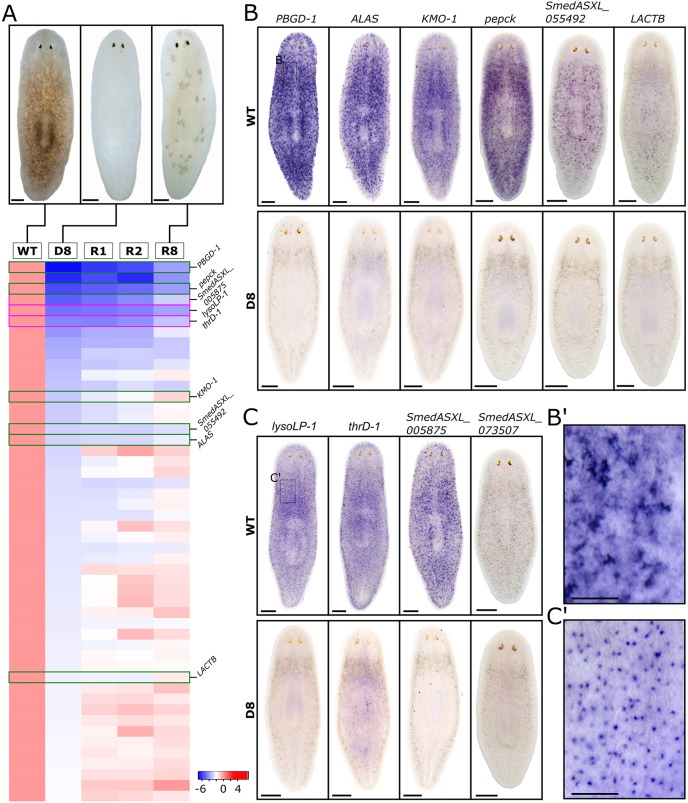
Fig. 1.
Identification of two classes of molecular markers for planarian pigment cells. (A) Whole-worm RNA samples were collected at five time points: before light exposure (‘WT’); exposed to light treatment for 8 days (D8); recovered in darkness for 1 day (R1), 2 days (R2) or 8 days (R8). (Top) Bright-field images of animal pigmentation status at time points WT, D8 and R8. Animals were fully depigmented at D8, whereas animals were partially repigmented at R8. (Bottom) Expression profile of 50 genes with the greatest downregulation at D8, in descending order of fold decrease. (B,C) WISH of candidate pigment cell markers. Six genes show dendritic expression patterns (B) and four genes show punctate expression patterns (C) by WISH. Top rows show that dendritic genes have varying degrees of expression in the subepidermal layer, whereas punctate genes have an even distribution across the animal in the subepidermal layer. Bottom rows show that gene expression is undetectable by WISH in depigmented animals at D8. (B′) Higher magnification image of neck region (boxed region in B), showing individual cells with dendritic expression of PBGD-1. (C′) Higher magnification image of neck region (boxed region in C), showing individual cells with punctate expression of lysoLP-1. Scale bars: 250 μm in A,B,C; 100 μm in B′,C′.
The first class revealed the dendritic morphology of pigment cells, and included the known pigment biosynthesis enzymes ALAS, PBGD-1 and KMO-1 (Stubenhaus et al., 2016). Light-induced loss of these markers was confirmed by WISH (Fig. 1B,B′). Two of the remaining three dendritic class genes had strong homology to the enzymes phosphoenolpyruvate carboxykinase (Smed-pepck) and β-lactamase (Smed-lactb). The second class of markers exhibited a more punctate WISH pattern that was more evenly distributed across the animal than for the dendritic class (Fig. 1C,C′). Cells expressing punctate markers clearly outnumbered cells expressing dendritic markers, but they were also ablated by light exposure, even more rapidly than dendritic cells (Fig. 1C, Fig. S2). Two of the four punctate markers had orthology to a 60 kDa lysophospholipase [Smed-lysoLP-1 (Sugimoto et al., 1998)] and a threonine dehydratase II (Smed-thrD-1). The mammalian ortholog of Smed-lysoLP-1 is mainly expressed in liver and kidney tissue (endoderm), and plays crucial roles in the hydrolysis and transacylation of multiple phosphatidylcholine derivatives (Sugimoto et al., 1998). The remaining transcripts did not exhibit detectable homology and were named from their transcript numbers (Fig. 1B,C). In total, we identified ten markers potentially defining two different subpopulations of light-sensitive pigment cells.
Candidate pigment cell subtypes partially overlap in gene expression and are localized to the muscle cell layer
We confirmed previous observations that KMO-1 and PBGD-1 are expressed in the same cells (Stubenhaus et al., 2016), and also determined that all dendritic markers were coincident by double fluorescent in situ hybridization (dFISH) (Fig. 2A). Similarly, we observed near complete overlap between different punctate class markers (Fig. 2A). Notably, FISH revealed that the punctate marker SmedASXL_005875 was expressed in cells with a dendritic morphology not evident by colorimetric WISH (Fig. 2A). Interestingly, 37±7% of cells expressing punctate markers also expressed dendritic markers, whereas 92±5% of cells expressing dendritic markers co-expressed punctate markers. We concluded that dendritic and punctate markers reflect two distinct cell types or states (see the Discussion), yet their overlapping expression also suggested that they might define cells in the same lineage.
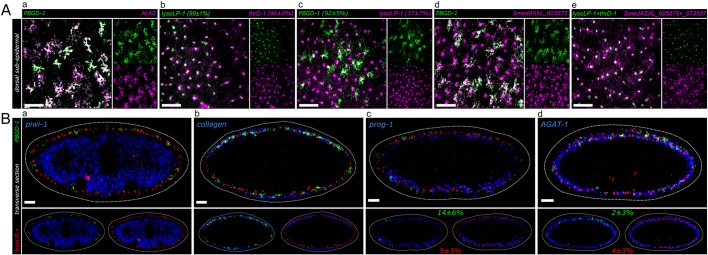
Fig. 2.
Co-expression analysis of pigment cell markers. (A) z-projections of coronal planes across 20 µm in the dorsal subepidermal layer by FISH. Co-expression is represented as follows: a percentage following a gene name indicates the percentage of cells labeled with this gene which also express the other gene in the same panel. (a) The dendritic markers PBGD-1 and ALAS show a high level of co-expression. (b) The punctate markers lysoLP-1 and thrD-1 show a high level of co-expression. (c) Dendritic+ cells (PBGD-1) highly co-express punctate markers, while a smaller subset of punctate+ cells (lysoLP-1) co-express dendritic markers. (d) Punctate marker-labeled cells possess dendritic cellular morphology. SmedASXL_005875 in situ signals can be detected in the processes of its marked cells. (e) Punctate markers with strict punctate cellular expression patterns (lysoLP-1 and thrD-1) are expressed in the same population that is labeled by punctate markers with process expression (SmedASXL_005875 and SmedASXL_073507). (B) Transverse sections of FISH indicate the localization of pigment cells (marked by PBGD-1 and lysoLP-1) in the subepidermal layer. (a) Pigment cells are located more distal-lateral to the stem cell compartment marked by piwi-1. (b) Pigment cells are interspersed with muscle cells labeled by collagen (SmedASXL_012760). (c) Pigment cells are more distal-lateral to early epidermal progenitor cells marked by prog-1. A small population of dendritic+ cells (14±6%) and punctate+ cells (9±3%) also expresses prog-1. (d) Pigment cells are in the same layer as late epidermal progenitor cells marked by AGAT-1. Scale bars: 50 μm.
We next analyzed the spatial location of dendritic and punctate markers in transverse sections using known markers for stem cells (piwi-1), epidermal progenitors (prog-1 and AGAT-1), and muscle (collagen) (Eisenhoffer et al., 2008; Reddien et al., 2005; Witchley et al., 2013; Zhu et al., 2015). Both dendritic and punctate markers were subepidermal, and interdigitated with muscle and epidermal progenitor cells (Fig. 2B), as predicted by previous electron microscopy studies of pigment cells in a related freshwater planarian species (Palladini et al., 1979). Interestingly, we observed a small but significant overlap between punctate and dendritic markers and early (prog-1+) and late (AGAT-1+) epidermal progenitors (between ∼2% and 14%; Fig. 2B), suggesting that the lineal origin of the pigment cells might be epithelial stem cells (also known as zeta-neoblasts) (van Wolfswinkel et al., 2014). However, when we ablated zeta-neoblasts using zfp-1(RNAi), we did not observe effects on either class of pigment cell markers (Fig. S3; see Discussion).
Dynamics and birth order of pigment cell markers during regeneration
Following head or tail amputation, regenerating tissue (the blastema) is initially unpigmented for 10-20 days before eventually developing full pigmentation. This allowed us to monitor the birth and/or migration of new pigment cells. To assay the dynamics of pigment cell markers in the blastema, trunk fragments regenerating new heads and tails were labeled for dendritic and punctate markers for 21 days post-amputation (Fig. 3A). Dendritic markers were detected in significantly higher numbers than punctate markers for the first ∼8 days of regeneration, and in lower numbers thereafter (Fig. 3A,B).
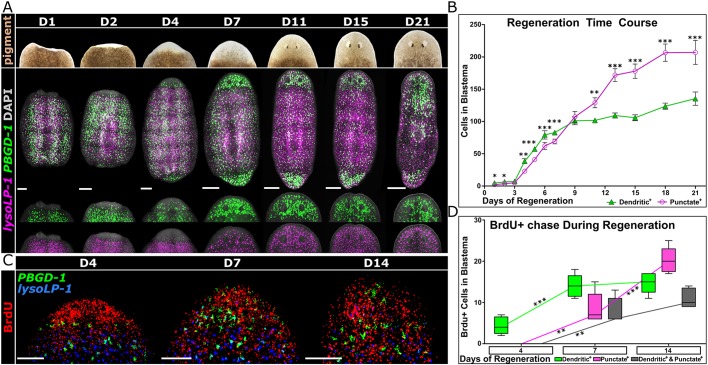
Fig. 3.
Distinct injury regeneration dynamics of dendritic+ versus punctate+ pigment cells. (A) (Top) Bright-field imaging of regenerating blastema in S. mediterranea from day 1 to day 21 post-head amputation, showing the repigmentation of head blastema. (Bottom) FISH of dendritic (PBGD-1) and punctate (lysoLP-1) markers during the same period, showing the restoration of the pigment cell population. (B) Regeneration dynamics of dendritic+ and punctate+ populations in head blastema as shown in A. Two-tailed, paired t-test was performed to determine the significance between dendritic+ and punctate+ populations within each time point. Error bars indicate s.d. (C) Birth order determination of dendritic+ and punctate+ cells by BrdU chase. BrdU was injected to trunk fragments 1 day after amputation. Animals were collected at 4, 7 and 14 days post-injection for BrdU detection, coupled with FISH of pigment cell markers PBGD-1 and lysoLP-1. (D) Birth order dynamics of dendritic+ and punctate+ cells as shown in C. Two-tailed, unpaired t-test was performed to determine the significance of variance between different time points for a certain population. Quantifications of the individual markers are inclusive of double-positive populations. *P<0.05, **P<0.01, ***P<0.001. Scale bars: 250 μm in A; 50 μm in C.
The emergence of dendritic and punctate markers in blastemas could be due to migration of pre-existing pigment cells. Alternatively, we reasoned that the predominance of dendritic markers at early stages of regeneration might reflect de novo formation of new pigment cells via a lineage in which cells expressing dendritic markers give rise to cells that express punctate markers. We tested these possibilities by birth-dating cells with the thymidine analog BrdU. Trunk fragments were injected with BrdU at 1 day post-amputation, and subsequently assayed for the appearance of dendritic and punctate markers as well as BrdU in head and tail blastemas (Fig. 3C). At day 4 of regeneration (3 days after the BrdU pulse), new pigment cells (BrdU+) were evident, and all of these expressed dendritic, but not punctate, markers (Fig. 3C,D). By day 7 of regeneration, BrdU was evident in cells expressing either punctate or dendritic markers, as well as cells co-expressing both markers (Fig. 3C,D). A similar trend continued through day 14, at which time the majority of the BrdU signal was in cells expressing punctate markers (Fig. 3D). These data show that new pigment cells are formed during regeneration, and are consistent with a lineage in which the newly born cells progress from expressing dendritic to punctate markers (see Discussion).
Dynamics and birth order of pigment cell markers during repigmentation
Similar to regeneration, animals that are fully depigmented after light exposure take ∼20-30 days to fully repigment (Fig. 4A) (Stubenhaus et al., 2016). We monitored the generation of new pigment cells in this context by dFISH with dendritic and punctate markers (Fig. 4A). As early as day 3 of repigmentation, increases in both classes of markers were evident (relative to residual pigment cells at day 0; Fig. S3B). In these new cells, we observed a very high degree of co-expression, peaking at >80% at day 5 of repigmentation (Fig. 4A). Thus, the earliest-born pigment cells express dendritic markers, as was observed during regeneration. In order to determine how pigment class markers correlate with the actual production of visual pigment, we performed single-worm dFISH combined with ʻlive' imaging immediately following fixation (Fig. 4B). Visual pigment cells in live images were circled and directly mapped to the same worm following dFISH for dendritic and punctate markers. We observed that until day 7 of repigmentation, all cells that were pigmented express dendritic markers. It was not until after day 11 that we observed any significant numbers of pigment+ punctate+ dendritic− cells. These data suggested that, as pigment cells mature and produce high levels of pigment, they downregulate dendritic markers of the pigment synthesis pathway.
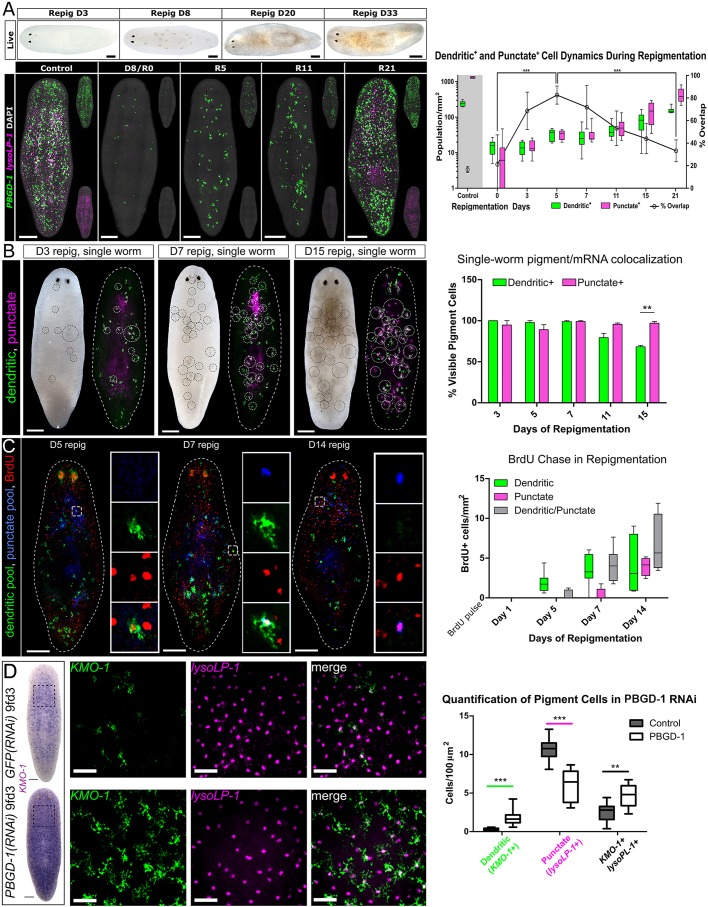
Fig. 4.
Dynamics and birth order of dendritic+ and punctate+ pigment cells during repigmentation. (A) (Top) Bright-field images showing repigmentation timeline of S. mediterranea following complete depigmentation by light exposure treatment. (Bottom) FISH of dendritic+ (PBGD-1) and punctate+ (lysoLP-1) pigment cells from repigmentation day 0 (R0) to day 21 (R21). (Right) Dendritic+ and punctate+ pigment cells demonstrate distinct birth dynamics after light-induced depigmentation. Percentage overlap refers to punctate+ cells that also express dendritic markers. Statistics shown: P-value (two-tailed, unpaired t-test) summary of percentage overlap comparisons between D0 and D5, and between D5 and D21. (B) Single-worm repigmentation live images combined with dFISH. An image was taken immediately following fixation and the pigmentation circled (left of each pair). Images were processed by changing the gamma value to enhance visualization of pigment. After dFISH for dendritic pooled markers and punctate pooled markers, the same circles were overlaid (right of each pair) and cells quantified inside the circles (bar chart on right). (C) (Left) BrdU pulse at day 0 of repigmentation and chased for the time shown. The boxed region is magnified in insets, which show single channels. (Right) Quantification of the identity of BrdU+ cells. (D) (Left) Representative animals stained for the dendritic marker KMO-1 in control (top row) or PBGD-1 (bottom row) RNAi. The boxed region is magnified in insets, which show dFISH for KMO-1 and lysoLP-1. (Right) Quantification of how markers change in PBGD-1 RNAi. Two-tailed, unpaired t-test. **P<0.01, ***P<0.001. Scale bars: 250 μm in A-D; 50 μm in boxed regions of D.
To further understand the timing of birth and differentiation of pigment cells during repigmentation, we injected animals at the end of depigmentation (day 0 of repigmentation) and monitored the chase of the label into each class of pigment cell marker. Similar to pigment cell regeneration, dendritic marker+ cells were labeled with BrdU first by 5 days, with a subsequent chase into dendritic+ punctate+ cells by 7 days and punctate+-only cells by 14 days (Fig. 4C). However, considerable overlap with both classes of markers was seen throughout, suggesting a continuum of differentiation.
Both regeneration and repigmentation assays demonstrated that dendritic+ cells precede punctate+ cells and might constitute a progenitor for mature (punctate) pigment cells. Such a ‘progenitor state’ would be consistent with other cell lineages in planarians, such as the gut, brain, muscle, protonephridia, eyes, and epidermis, all of which have a postmitotic progenitor-like cell state en route to final differentiated cell types (Currie et al., 2016b; Eisenhoffer et al., 2008; Forsthoefel et al., 2012; Lapan and Reddien, 2012; Scimone et al., 2014, 2011; Witchley et al., 2013; Zhu et al., 2015). For these other lineages (particularly the epithelium), stem cell ablation using gamma-irradiation (Dubois and Wolff, 1947; Hayashi et al., 2006) leads to progenitor loss several days later (Eisenhoffer et al., 2008; Zhu et al., 2015). Thus, we reasoned that cells expressing dendritic markers might be irradiation sensitive if they define a progenitor for mature pigment cells. To test this hypothesis, we lethally irradiated fully pigmented animals and assayed changes in dendritic and punctate markers for 14 days (near the terminal point of survival for these animals). Surprisingly, we detected no changes in the number of cells expressing dendritic or punctate markers, demonstrating that there is little turnover of pigment cells during tissue homeostasis (Fig. S4). To determine whether expression of dendritic markers constitutes a progenitor state only when animals are challenged to make new pigment cells, we lethally irradiated animals 8 days after ablating pigment cells by light exposure. In these repigmenting animals, the number of cells expressing punctate markers remained constant, whereas the number of cells expressing dendritic markers significantly decreased (Fig. S5). These data suggested that dendritic markers might define an immature progenitor within the pigment cell lineage, at least when animals are forced to generate new pigment cells.
An alternative to dendritic+ cells being progenitors is that cells can dynamically switch between cell states (see Discussion). To test whether dendritic markers can be dynamically controlled, we performed RNAi to PBGD-1, which results in white animals without the loss of pigment cells (Stubenhaus et al., 2016). As animals became whiter, we observed significant increases in the numbers of cells expressing the dendritic marker KMO-1, as well as in the number of dendritic+ punctate+ cells (Fig. 4D). Combined with a concomitant decrease in punctate+-only cells, this suggested that pigment class markers can be dynamically controlled.
Identification of regulators of the pigment cell lineage
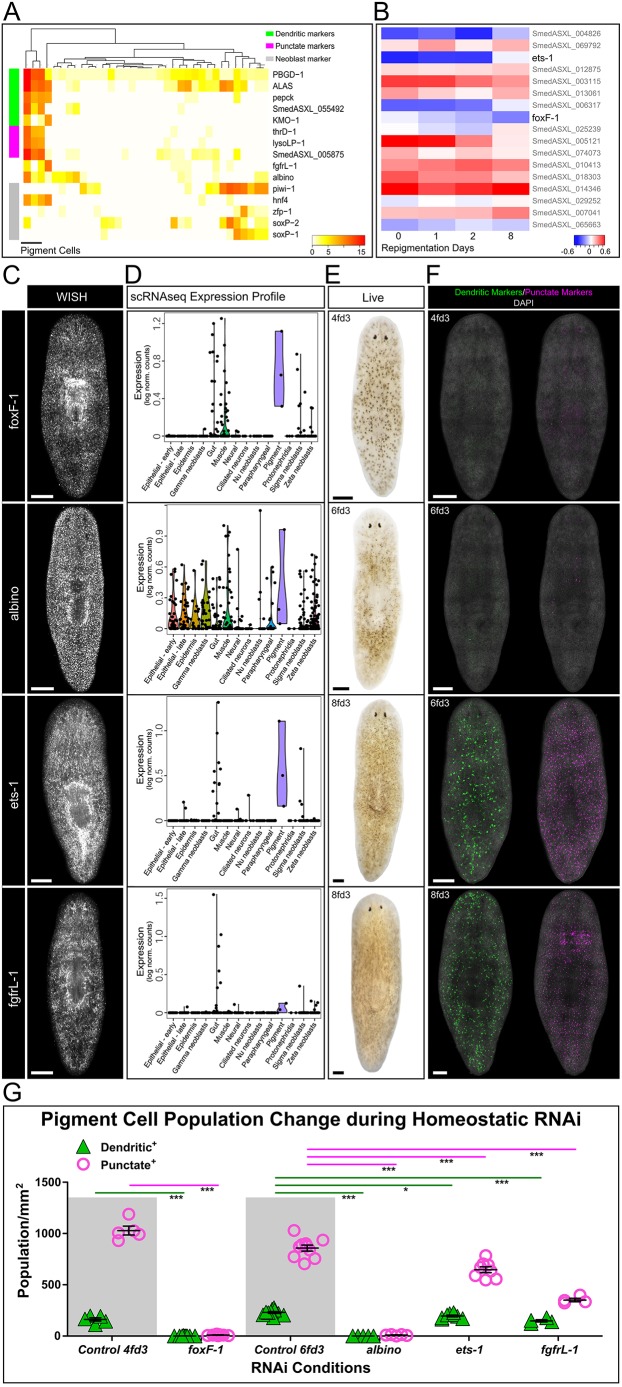
Using available single-cell RNAseq (scRNAseq) datasets (Molinaro and Pearson, 2016; Wurtzel et al., 2015), we identified 35 cells with detectable expression of PBGD-1. RNAi knockdown of this gene produces white animals (Stubenhaus et al., 2016). When the PBGD-1+ cells were hierarchically clustered, a cluster of three cells stood out as being likely pigment cells in that they expressed all dendritic and punctate markers (Fig. 5A; see Discussion). Importantly, the three identified pigment cells also co-expressed the known pigment cell regulator and forkhead transcription factor albino (Wang et al., 2016) and an FGF receptor (FGFR)-like molecule, Smed-fgfrL-1, also required for normal pigmentation (Dr Francesc Cebria, personal communication; transcript number SmedASXL_006982). These three cells were previously categorized as ʻgut' (Wurtzel et al., 2015), and together with the observation that lysoLP-1 and thrD-1 are liver-enriched genes in mammals (Leoncini et al., 1998; Sugimoto et al., 1998), this could suggest that pigment cells may have an endodermal origin. In addition, all three pigment cells highly express the putative endodermal-specific ʻgamma-neoblast' marker hnf4, which indicated that planarian pigment cells might originate from stem cells with endodermal characteristics (Martín-Durán and Romero, 2011; van Wolfswinkel et al., 2014). We tested this idea by RNAi knockdown of hnf4, and although the experiment was complicated by the health defects that occur when RNAi animals are also exposed to light, we did observe some changes in the intensity of dendritic marker staining (Fig. S6). Thus, it is interesting to speculate that planarian pigment cells have endodermal, rather than ectodermal, origins (see Discussion).
Fig. 5.
Identification of pigment cell lineage regulators by scRNAseq. (A) Expression analysis of pigment cell markers and known lineage regulators in 35 PBGD-1+ single cells. Neoblast markers are shown at the bottom. (B) Expression analysis of the 17 transcription factors that have a >0.5 correlation with pigment cell marker (PBGD-1) expression. (C) WISH of transcription factors foxF-1, ets-1 and albino and of the FGFR-like molecule fgfrL-1 reveals broad expression patterns in S. mediterranea. (D) Single-cell expression analysis indicates high levels of foxF-1, ets-1 and fgfrL-1 expression in pigment cells. albino shows high expression in pigment cells but is also broadly expressed in other tissue types. (E) Bright-field imaging of homeostatic pigmentation phenotype after RNAi of foxF-1, ets-1, albino and fgfrL-1. foxF-1 and albino RNAi lead to more severe depigmentation than ets-1 and fgfrL-1 RNAi. For time point nomenclature, see Materials and Methods. Refer to Fig. 1A for control pigmentation. (F) dFISH images of pooled dendritic (PBGD-1 and ALAS) and punctate (lysoLP-1 and thrD-1) markers after candidate pigment lineage regulator RNAi. (G) Quantification of dendritic+ and punctate+ population densities at the end of foxF-1, ets-1, albino and fgfrL-1 RNAi treatment. *P<0.05, **P<0.01, ***P<0.001. Two-tailed, unpaired t-test. Scale bars: 250 μm.
We next analyzed the co-expression of transcription factors that were highly correlated with PBGD-1 in the scRNAseq data (Pearson correlation ≥0.3) (Fig. 5B). In total, we found 17 transcription factors that met this criterion (Table S2). We performed an RNAi screen of the transcription factors co-expressed in the pigment cells, and found two that were required for normal pigmentation pattern or intensity of pigmentation, similar to Smed-fgfrL-1 and albino: a FoxF homolog (Smed-foxF-1) and an ETS-1 family homolog (Smed-ets-1) (Fig. 5C-G).
foxF-1 and ets-1 have differential requirements in pigment cells
We assayed RNAi knockdown of the above genes for alterations of both dendritic and punctate markers in uninjured animals using pooled dFISH probes. Following knockdown of either albino or foxF-1, both punctate and dendritic markers were virtually ablated (Fig. 5F,G). fgfrL-1(RNAi) animals also showed reduction, but not complete ablation, of these markers. By contrast, ets-1 knockdown predominantly affected cells expressing punctate markers (Fig. 5F,G). It should be noted that RNAi of each factor was eventually lethal to the animals, with the exception of albino; however, we did not observe overt defects in other known cell types outside of the pigment lineage (Fig. S7).
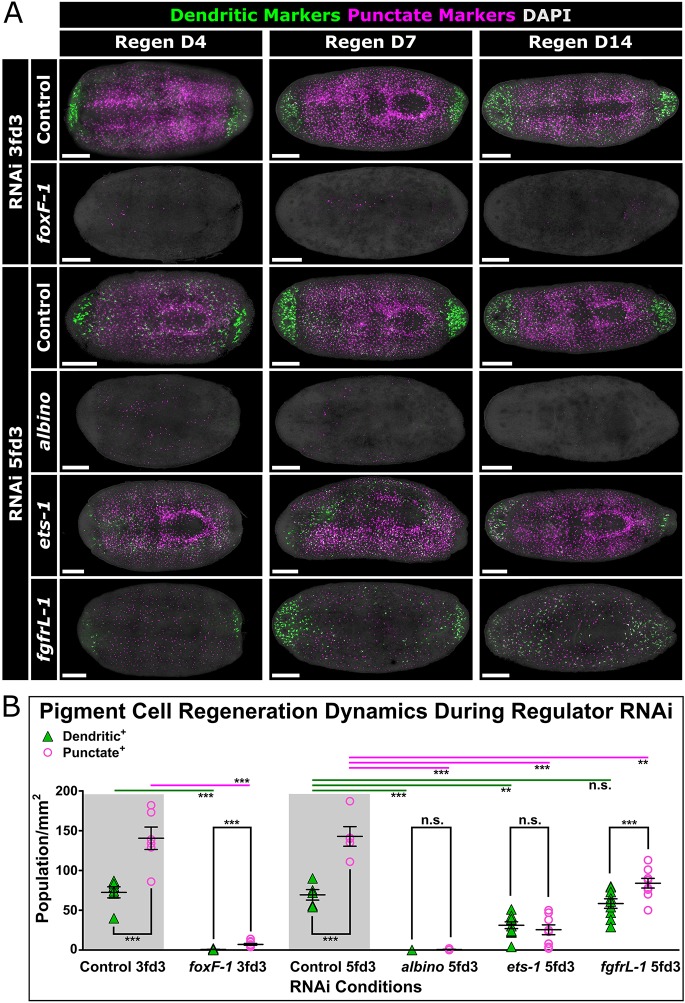
These RNAi experiments were performed on steady-state animals, where we have shown the relationships between dendritic and punctate markers to be somewhat different than when animals are challenged to remake pigment cells de novo (Fig. 4). Therefore, we next tested the role of each factor in the regeneration of new pigment cells. Both foxF-1(RNAi) and albino(RNAi) animals showed total ablation of both dendritic and punctate markers during regeneration (Fig. 6A). ets-1(RNAi) animals, on the other hand, showed reduced numbers of cells expressing both marker classes compared with controls (Fig. 6A,B). We conclude that ets-1 functions to maintain the punctate cell state in homeostatic animals, but is required for pigment cell production during regeneration. fgfrL-1(RNAi) animals showed more modest, yet measurable, defects in the regeneration of punctate markers only (Fig. 6A,B).
Fig. 6.
foxF-1, albino, ets-1 and fgfrL-1 regulate the pigment cell lineage during regeneration. (A) FISH with pooled RNA probes of dendritic and punctate markers on regenerating RNAi animals. Animals were amputated after the indicated RNAi feedings (e.g. amputated 3 days after the last feeding for foxF-1 RNAi experiment) and imaged at 4, 7 and 14 days after amputation. (B) Quantification of dendritic+ and punctate+ cells at D14 of regeneration during RNAi of pigment cell regulators. Statistics: top, two-tailed, unpaired t-test; within each RNAi condition, two-tailed, paired t-test. **P<0.01, ***P<0.001; n.s., not significant. Scale bars: 250 μm.
In summary, we have identified multiple transcriptional regulators of pigment cells, as well as a potential signaling pathway involving an FGFR-like molecule that is likely to be involved in pigment cell specification and differentiation.
DISCUSSION
Our characterization of gene expression profiles in S. mediterranea pigment cells provides new insight into the developmental origins of body pigmentation in planarians. Specifically, we have identified a series of molecular markers for the pigment cell lineage and shown that production and maintenance of pigment cells during regeneration and tissue homeostasis is controlled by an FGFR-like molecule and the FOXF-1 and ETS-1 transcription factors.
Dynamics of gene expression within the pigment cell lineage
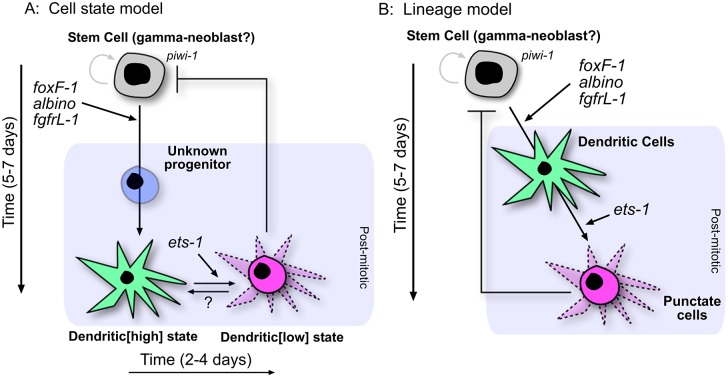
We identified and characterized two classes of pigment cell markers exhibiting spatial and temporal differences in expression (Figs 1-4). These data are consistent with two different models for the pigment cell lineage: (1) all pigment cells are equivalent and capable of expressing dendritic markers, punctate markers, or both in response to developmental cues (Fig. 7A); (2) cells expressing dendritic markers are precursors for terminally differentiated pigment cells, which express only punctate markers (Fig. 7B). We favor the former model for several reasons. First, previous electron microscopy studies reported that mature pigment cells have a dendritic morphology in planarians (Palladini et al., 1979), as in many other animal species. Second, although there are clear differences in the number of cells expressing dendritic versus punctate markers during physiological cell turnover in adult animals (at the resolution of WISH), these markers show significant overlap (Fig. 2). Furthermore, FISH revealed that at least one punctate marker (SmedASXL_005875) was expressed in cells exhibiting a dendritic morphology (Fig. 2A), suggesting that the punctate expression pattern might reflect confinement of mRNAs to the cell body, rather than a difference in cell morphology. Third, scRNAseq revealed co-expression of all dendritic and punctate markers in all three pigment cells analyzed (Fig. 5A). Finally, RNAi knockdown of the dendritic markers KMO-1 or PBGD-1 uniformly alters or eliminates normal bodily pigmentation (Stubenhaus et al., 2016), rather than leading to a salt-and-pepper appearance as might be expected if only a fraction of pigment cells was affected.
Fig. 7.
Two primary models of planarian pigment cell development. (A) Cell state model during pigment cell homeostasis. This model suggests that dendritic+ and punctate+ cells are all pigment cells that can dynamically switch between cell states, but that these states are essentially equivalent. We cannot account for pigment cells prior to turning on dendritic markers, which takes ∼5 days (Fig. 4C). We propose there is an unknown progenitor state to account for this time gap. During repigmentation, it takes an addition 2-4 days for punctate+-only cells to be seen (Fig. 4C). (B) Because all early-born pigment cells are dendritic marker+ during regeneration and repigmentation (Figs 3, 4) and the end point of the fully pigmented cell is punctate+-only, the lineage might not be dynamic in marker expression and differentiation is one way, with dendritic+ cells as a progenitor. It should be noted that RNAi of each regulatory factor has slightly different effects on the lineage during regeneration (Fig. 6) as opposed to what is shown here for homeostasis.
We found that cells expressing dendritic markers are the first to appear during both the pigmentation of newly regenerated tissue (Fig. 3) and the repigmentation of depigmented tissue (Fig. 4A-C). Although these observations are formally consistent with a dendritic precursor model (Fig. 7B), we instead propose that they reflect upregulation of pigment biosynthesis pathways necessary for ommochrome and porphyrin production in new pigment cells. Indeed, three of the six dendritic markers we identified (but none of the punctate markers) are known pigment biosynthesis enzymes [ALAS, PBGD-1 and KMO-1 (Stubenhaus et al., 2016)]. Once new pigment granules have been synthesized, expression of these enzymes might be downregulated in concert with upregulation of punctate class genes. The functional significance of this switch awaits further characterization of the punctate markers, but it is interesting to note that it might be reversible, as we observed a dramatic upregulation of dendritic class markers when pigment biosynthesis was inhibited via RNAi knockdown of PBGD-1 (Fig. 4D).
What is the stem cell of origin for planarian pigment cells?
dFISH with dendritic or punctate markers together with markers for progenitors of the epithelial lineage revealed a low but significant overlap in expression (particularly for prog-1+ ʻearly progenitors'; Fig. 2B). Because the epithelial lineage appears to have a dedicated stem cell class, i.e. the zeta-neoblast (van Wolfswinkel et al., 2014), we tested whether zfp-1(RNAi) (zeta-neoblast-ablated) animals could make new pigment cells. They could (Fig. S3), suggesting that the pigment cell lineage is not downstream of zeta-neoblasts [although these experiments were complicated by the poor health of zfp-1(RNAi) animals, which was compounded by depigmentation/amputation].
Unlike RNAi knockdown of zfp-1, RNAi knockdown of hnf4 did affect the staining intensity of dendritic cells during repigmentation and regeneration (Fig. S6). Moreover, lysoLP-1, thrD-1 and hnf4 were strongly expressed in all three pigment cells analyzed by scRNAseq (Fig. 5A). This raises the interesting possibility that the pigment cell lineage might overlap with the lineage of endodermal tissues such as the gut. It will be interesting to test this hypothesis in the future, when endodermal neoblasts are better defined.
By analyzing irradiated animals, we determined that pigment cells do not exhibit significant turnover during the course of the 2 weeks that animals can be assayed (Fig. S4). This implies that, once an animal is fully pigmented, stem cells no longer drive more than a very low rate of new pigment cell production. In turn, this might suggest a feedback loop in which mature pigment cells repress pigment cell specification (Fig. 7). The existence of such a regulatory mechanism might explain the variable RNAi phenotypes observed for fgfrL-1 and ets-1 during tissue homeostasis and regeneration (Figs 5, 6).
Factors controlling pigment cell homeostasis and patterning
Our data also provide insight into the molecular mechanisms regulating the pigment cell lineage. The FGFR-like molecule encoded by fgfrL-1 is required for normal pigment cell specification (Figs 5, 6), similar to the function of a homologous pathway in the invertebrate chordate Ciona (Curran et al., 2009; Racioppi et al., 2014; Squarzoni et al., 2011; Thomas and Erickson, 2009). The identity and source of the ligand for this receptor are presently unknown, but it is interesting to speculate that it might be secreted by muscle, both because the pigment cells are interdigitated with the muscle layer (Fig. 2) (Palladini et al., 1979) and because many other patterning signals are secreted by muscle cells in planarians (Lander and Petersen, 2016; Scimone et al., 2016; Witchley et al., 2013).
RNAi knockdown of foxF-1 led to complete ablation of both dendritic and punctate class markers during tissue homeostasis and regeneration (Figs 5, 6), suggesting that this transcription factor is a key regulator of pigment cell production. The related family member Foxd3 controls melanophore specification in zebrafish and chick by regulating expression of the microphthalmia-associated transcription factor (Mitf) (Curran et al., 2009; Racioppi et al., 2014; Squarzoni et al., 2011; Thomas and Erickson, 2009). There are multiple Mitf homologs in S. mediterranea EST databases, raising the possibility that this regulatory mechanism is conserved in planarians. RNAi knockdown of the transcription factors albino and ets-1 also affected pigment cell specification, with albino showing a strong RNAi phenotype like foxF-1 (Figs 5, 6) (Wang et al., 2016), and ets-1(RNAi) predominantly affecting the expression of punctate markers, like fgfrL-1 knockdown during regeneration (Figs 5, 6). Identifying the regulatory hierarchy of these genes represents a key goal for future studies.
What is the function of pigment cells in planarian biology (and what about cave planarians)?
Pigment cells function in processes as diverse as mating, mimicry, camouflage, and adaptation to UV, all of which are subject to strong selective forces (sexual and natural). In planarians, the body pigment cells may be involved in the sensing of light [the ʻnon-ocular light response' (Birkholz and Beane, 2017)], as well as camouflage from predators, although neither has been rigorously tested. In our studies, animals that survived the depigmentation process appeared perfectly normal, suggesting that there is no essential function for pigment cells in S. mediterranea (Stubenhaus et al., 2016).
As observed in many other animal lineages, cave-dwelling planarians have lost body pigmentation, further supporting the notion that pigment cells are dispensable for core biological functions. It remains unknown whether this reflects loss of pigment cells or pigment biosynthesis enzymes (Hyman, 1951). Thus, it will be interesting in future studies to characterize the evolutionary mechanisms leading to the loss of pigment cells or biosynthetic pathways in various cave planarian species by conducting a careful examination of the markers and regulators that we have reported here.
MATERIALS AND METHODS
Animal husbandry, exposure to γ-irradiation, and depigmentation
Asexual Schmidtea mediterranea strain CIW4 were reared as previously described (Sánchez Alvarado, 2002). For irradiation experiments, planarians were exposed to 60 Gray (Gy) of γ-irradiation from a 137Cs source (Pearson and Sánchez Alvarado, 2010). For depigmentation experiments, planarians were exposed to light at intermittent intervals as previously described (Stubenhaus et al., 2016). Animals were exposed to white light from LED light strips for 15 min, followed by 225 min of recovery in the dark. This cycle was repeated six times per day for 8-11 days, or until animals had lost all visible body pigment. A fan was used to prevent temperature elevation during light exposure, and animals were maintained in 50 µg/ml gentamycin and rinsed daily throughout the process. Size-matched control animals were kept in the dark in a 20°C incubator. Following depigmentation, animals were kept in the dark in an incubator for up to 33 days to allow for repigmentation. Planarians were prepared for bright-field imaging by incubation in 2% HCl for 15 s, then rinsing with PBS. Animals were immediately imaged using a Leica M165 fluorescence dissecting microscope.
RNAi
RNAi experiments were performed using previously described expression constructs and E. coli HT115 (Newmark et al., 2003). Bacteria were grown to an OD600 of 0.8 and induced with 1 mM IPTG for 2 h. Bacteria were pelleted, mixed with liver paste at a ratio of 333 µl liver to 100 ml original culture volume, and frozen in aliquots. This dilution was used in all experiments except for the zfp-1 knockdown repigmentation experiment, in which 500 µl liver was mixed with 100 ml original culture volume. The negative control, ʻcontrol(RNAi)', was the gfp sequence as previously described (Cowles et al., 2013). RNAi food was fed to 1-week-starved experimental worms twice weekly for a total of two to eight feedings. The total number of feedings, as well as the number of days after the last feeding, are denoted in each figure; for example, 6fd3 indicates six feedings and 3 days since the final feeding. Amputations were performed 3 days after the final feeding, with only the trunk fragment used in experiments. For the combination RNAi and repigmentation experiments, animals were fed twice during depigmentation: zfp-1(RNAi) animals were fed on day 0 and day 3 of depigmentation, whereas hnf-4(RNAi) animals were fed on day 5 and day 8. All animals were size-matched between experimental and control worms.
Immunolabeling, BrdU, and in situ hybridization (ISH)
Whole-mount ISH (WISH), double fluorescent ISH (dFISH), and immunostainings were performed as previously described (Currie et al., 2016a; Lauter et al., 2011; Pearson et al., 2009). Colorimetric WISH stains were imaged on a Leica M165 fluorescence dissecting microscope. dFISH and BrdU stains were imaged on a Leica DMIRE2 inverted fluorescence microscope with a Hamamatsu back-thinned EM-CCD camera and spinning disc confocal scan head. BrdU (Sigma, B5002-5G; 25 mg/ml) was dissolved at 25 mg/ml in 50% ethanol and injected into the gut of each planarian using a Nanoject II microinjector (Drummond) and stained as previously described (Zhu et al., 2015). Planarians for BrdU experiments were maintained in a high salt concentration [2.5 g/l of Instant Ocean (∼43 mM)] beginning 1 week prior to injection. For regeneration experiments, trunk fragments were injected with BrdU 1 day after amputation of heads and tails. Cell counts and colocalizations were quantified using the Cell Counter function in ImageJ (http://rsb.info.nih.gov/ij/). Positive cells were visually distinguished manually. Significance was determined by a two-tailed, unequal variance, pairwise Student's t-test. Images were post-processed in a similar manner using Adobe Photoshop. Brightfield images of live worms in Fig. 4B were processed by changing the gamma to enhance visualization of pigment.
RNAseq, co-expression, and differential expression analysis
RNA deep sequencing (RNAseq) was performed on wild-type (WT) animals as well as fully depigmented animals (8 days of depigmentation by light exposure) and partially repigmented animals (1, 2 and 8 days of repigmentation in the dark on starved animals). Experiments were performed in biological triplicate, sequenced to a depth of ∼40 million reads per sample, and multiplexed on an Illumina HiSeq2500 with 50 bp, single-end reads. Seventy-five genes of interest were identified based on high 8 day WT versus depigmented fold change (Table S1). Raw scRNAseq data (including stem cells, neurons, gut, epithelial, muscle and parapharyngeal cells) were obtained from NCBI Sequence Read Archive (PRJNA276084) and Gene Expression Omnibus (GSE79866) (Molinaro and Pearson, 2016; Wurtzel et al., 2015). Reads were aligned to the SmedASXL transcriptome assembly under NCBI BioProject PRJNA215411 using bowtie2 (Langmead and Salzberg, 2012) with 15 bp 3′ trimming. Violin plots were produced using modified source code from Macosko et al. (2015) and heatmaps were produced using the modified heatmap.3 source code from Molinaro and Pearson (2016). Heatmaps in Fig. 1A and Fig. 5B have expression levels row-normalized to WT values for ease of visualization. The heatmap in Fig. 5A has log2 normalized expression counts. Pigment lineage transcription factors were identified based on expression correlation with PBGD-1 (Pearson correlation ≥0.3; Table S2) in the published scRNAseq data (Molinaro and Pearson, 2016; Wurtzel et al., 2015).
Supplementary Material
Acknowledgements
We thank Dr Francesc Cebria for sharing fgfrL-1 information (transcript sequence and phenotype); and Dr Jochen Rink for sharing anti-PIWI-1 antibody.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: X.H., N.L.-M., A.M.M., J.P., B.J.P.; Methodology: X.H., N.L.-M., A.M.M., J.P., B.J.P.; Software: A.M.M.; Validation: X.H., N.L.-M., Y.L.; Formal analysis: X.H., N.L.-M., Y.L.; Investigation: B.J.P.; Resources: B.J.P.; Data curation: X.H., N.L.-M., Y.L., A.M.M.; Writing - original draft: B.J.P.; Writing - review & editing: X.H., N.L.-M., Y.L., A.M.M., J.P.; Supervision: J.P., B.J.P.; Project administration: B.J.P.; Funding acquisition: J.P., B.J.P.
Funding
N.L.-M. was funded by a Restracomp student fellowship from the Hospital for Sick Children. X.H. was funded by Natural Sciences and Engineering Research Council of Canada (NSERC) operating grant RGPIN-2016-06354. A.M.M. was funded by Canadian Institutes of Health Research Banting and Best PhD fellowship GSD-152379. B.J.P. was funded by Ontario Institute for Cancer Research Investigator Award IA-026. J.P. was funded by grants from the National Institutes of Health (1R15GM107826-01) and National Science Foundation (IOS-1445541). Deposited in PMC for release after 12 months.
Data availability
Raw RNAseq data and bowtie2 outputs from this manuscript are available at NCBI Gene Expression Omnibus (GEO) under accession number GSE106219.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.156349.supplemental
References
- Adameyko I., Lallemend F., Aquino J. B., Pereira J. A., Topilko P., Müller T., Fritz N., Beljajeva A., Mochii M., Liste I. et al. (2009). Schwann cell precursors from nerve innervation are a cellular origin of melanocytes in skin. Cell 139, 366-379. 10.1016/j.cell.2009.07.049 [DOI] [PubMed] [Google Scholar]
- Aspengren S., Hedberg D., Sköld H. N. and Wallin M. (2009). New insights into melanosome transport in vertebrate pigment cells. Int. Rev. Cell Mol. Biol. 272, 245-302. 10.1016/S1937-6448(08)01606-7 [DOI] [PubMed] [Google Scholar]
- Birkholz T. R. and Beane W. S. (2017). The planarian TRPA1 homolog mediates extraocular behavioral responses to near-ultraviolet light. J. Exp. Biol. 220, 2616-2625. 10.1242/jeb.152298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarelli-Neto O., Ferreira A. S., Martins W. K., Pavani C., Severino D., Faião-Flores F., Maria-Engler S. S., Aliprandini E., Martinez G. R., Di Mascio P. et al. (2014). Melanin photosensitization and the effect of visible light on epithelial cells. PLoS ONE 9, e113266 10.1371/journal.pone.0113266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichorek M., Wachulska M., Stasiewicz A. and Tymińska A. (2013). Skin melanocytes: biology and development. Postepy Dermatol. Alergol. 30, 30-41. 10.5114/pdia.2013.33376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles M. W., Brown D. D. R., Nisperos S. V., Stanley B. N., Pearson B. J. and Zayas R. M. (2013). Genome-wide analysis of the bHLH gene family in planarians identifies factors required for adult neurogenesis and neuronal regeneration. Development 140, 4691-4702. 10.1242/dev.098616 [DOI] [PubMed] [Google Scholar]
- Curran K., Raible D. W. and Lister J. A. (2009). Foxd3 controls melanophore specification in the zebrafish neural crest by regulation of Mitf. Dev. Biol. 332, 408-417. 10.1016/j.ydbio.2009.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie K. W., Brown D. D. R., Zhu S., Xu C. J., Voisin V., Bader G. D. and Pearson B. J. (2016a). HOX gene complement and expression in the planarian Schmidtea mediterranea. Evodevo 7, 7 10.1186/s13227-016-0044-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie K. W., Molinaro A. M. and Pearson B. J. (2016b). Neuronal sources of hedgehog modulate neurogenesis in the adult planarian brain. Elife 5, e19735 10.7554/eLife.19735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois F. and Wolff E. (1947). Sur une méthode d'irradiation localisée permettant de mettre en évidence la migration des cellules de régénération chez les planaires. C. R. Seances Soc. Biol. Fil. 141, 903-906. [PubMed] [Google Scholar]
- Eisenhoffer G. T., Kang H. and Sanchez Alvarado A. (2008). Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell 3, 327-339. 10.1016/j.stem.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom D. S., Bain E. J., Patterson L. B., Grout M. E. and Parichy D. M. (2015). Long-distance communication by specialized cellular projections during pigment pattern development and evolution. Elife 4, e12401 10.7554/eLife.12401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel D. J., James N. P., Escobar D. J., Stary J. M., Vieira A. P., Waters F. A. and Newmark P. A. (2012). An RNAi screen reveals intestinal regulators of branching morphogenesis, differentiation, and stem cell proliferation in planarians. Dev. Cell 23, 691-704. 10.1016/j.devcel.2012.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgeier M. H., Klein L. E., Klein-Angerer S., Moellmann G. and Nordlund J. J. (1984). The distribution of melanocytes in the leptomeninges of the human brain. J. Invest. Dermatol. 82, 235-238. 10.1111/1523-1747.ep12260111 [DOI] [PubMed] [Google Scholar]
- Hase S., Wakamatsu K., Fujimoto K., Inaba A., Kobayashi K., Matsumoto M., Hoshi M. and Negishi S. (2006). Characterization of the pigment produced by the planarian, Dugesia ryukyuensis. Pigment Cell Res. 19, 248-249. 10.1111/j.1600-0749.2006.00306.x [DOI] [PubMed] [Google Scholar]
- Hayashi T., Asami M., Higuchi S., Shibata N. and Agata K. (2006). Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev. Growth Differ. 48, 371-380. 10.1111/j.1440-169X.2006.00876.x [DOI] [PubMed] [Google Scholar]
- Hyman L. H. (1951). The Invertebrates Vol. 2: Platyhelminthes and Rhynchocoela, the Acoelomate Bilateria. New York: McGraw-Hill. [Google Scholar]
- Lander R. and Petersen C. P. (2016). Wnt, Ptk7, and FGFRL expression gradients control trunk positional identity in planarian regeneration. Elife 5, e12850 10.7554/eLife.12850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. and Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357-359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan S. W. and Reddien P. W. (2012). Transcriptome analysis of the planarian eye identifies ovo as a specific regulator of eye regeneration. Cell Rep. 2, 294-307. 10.1016/j.celrep.2012.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter G., Söll I. and Hauptmann G. (2011). Two-color fluorescent in situ hybridization in the embryonic zebrafish brain using differential detection systems. BMC Dev. Biol. 11, 43 10.1186/1471-213X-11-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoncini R., Vannoni D., Di Pietro M. C., Guerranti R., Rosi F., Pagani R. and Marinello E. (1998). Restoration of rat liver L-threonine dehydratase activity by pyridoxamine 5′-phosphate: the half-transaminating activity of L-threonine dehydratase and its regulatory role. Biochim. Biophys. Acta 1425, 411-418. 10.1016/S0304-4165(98)00094-4 [DOI] [PubMed] [Google Scholar]
- Lister J. A. (2002). Development of pigment cells in the zebrafish embryo. Microsc. Res. Tech. 58, 435-441. 10.1002/jemt.10161 [DOI] [PubMed] [Google Scholar]
- Macosko E. Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A. R., Kamitaki N., Martersteck E. M. et al. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202-1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Durán J. M. and Romero R. (2011). Evolutionary implications of morphogenesis and molecular patterning of the blind gut in the planarian Schmidtea polychroa. Dev. Biol. 352, 164-176. 10.1016/j.ydbio.2011.01.032 [DOI] [PubMed] [Google Scholar]
- Mills M. G. and Patterson L. B. (2009). Not just black and white: pigment pattern development and evolution in vertebrates. Semin. Cell Dev. Biol. 20, 72-81. 10.1016/j.semcdb.2008.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjaatvedt C. H., Kern C. B., Norris R. A., Fairey S. and Cave C. L. (2005). Normal distribution of melanocytes in the mouse heart. Anat. Rec. A Discov. Mol. Cell Evol. Biol. 285A, 748-757. 10.1002/ar.a.20210 [DOI] [PubMed] [Google Scholar]
- Molinaro A. M. and Pearson B. J. (2016). In silico lineage tracing through single cell transcriptomics identifies a neural stem cell population in planarians. Genome Biol. 17, 87 10.1186/s13059-016-0937-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark P. A. and Sanchez Alvarado A. (2002). Not your father's planarian: a classic model enters the era of functional genomics. Nat. Rev. Genet. 3, 210-219. 10.1038/nrg759 [DOI] [PubMed] [Google Scholar]
- Newmark P. A., Reddien P. W., Cebrià F. and Sánchez Alvarado A. (2003). Ingestion of bacterially expressed double-stranded RNA inhibits gene expression in planarians. Proc. Natl. Acad. Sci. USA 100 Suppl. 1, 11861-11865. 10.1073/pnas.1834205100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura E. K., Jordan S. A., Oshima H., Yoshida H., Osawa M., Moriyama M., Jackson I. J., Barrandon Y., Miyachi Y. and Nishikawa S.-I. (2002). Dominant role of the niche in melanocyte stem-cell fate determination. Nature 416, 854-860. 10.1038/416854a [DOI] [PubMed] [Google Scholar]
- Noonan F. P., Zaidi M. R., Wolnicka-Glubisz A., Anver M. R., Bahn J., Wielgus A., Cadet J., Douki T., Mouret S., Tucker M. A. et al. (2012). Melanoma induction by ultraviolet A but not ultraviolet B radiation requires melanin pigment. Nat. Commun. 3, 884 10.1038/ncomms1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladini G., Medolago-Albani L., Margotta V., Conforti A. and Carolei A. (1979). The pigmentary system of planaria. I. Morphology. Cell Tissue Res. 199, 197-202. 10.1007/BF00236131 [DOI] [PubMed] [Google Scholar]
- Parichy D. M. and Spiewak J. E. (2015). Origins of adult pigmentation: diversity in pigment stem cell lineages and implications for pattern evolution. Pigment Cell Melanoma Res. 28, 31-50. 10.1111/pcmr.12332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan W. J. and Raible D. W. (2012). Specification of neural crest into sensory neuron and melanocyte lineages. Dev. Biol. 366, 55-63. 10.1016/j.ydbio.2012.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B. J. and Sánchez Alvarado A. (2010). A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development 137, 213-221. 10.1242/dev.044297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B. J., Eisenhoffer G. T., Gurley K. A., Rink J. C., Miller D. E. and Sanchez Alvarado A. (2009). Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev. Dyn. 238, 443-450. 10.1002/dvdy.21849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premi S., Wallisch S., Mano C. M., Weiner A. B., Bacchiocchi A., Wakamatsu K., Bechara E. J., Halaban R., Douki T. and Brash D. E. (2015). Photochemistry. Chemiexcitation of melanin derivatives induces DNA photoproducts long after UV exposure. Science 347, 842-847. 10.1126/science.1256022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racioppi C., Kamal A. K., Razy-Krajka F., Gambardella G., Zanetti L., di Bernardo D., Sanges R., Christiaen L. A. and Ristoratore F. (2014). Fibroblast growth factor signalling controls nervous system patterning and pigment cell formation in Ciona intestinalis. Nat. Commun. 5, 4830 10.1038/ncomms5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls J. F. and Johnson S. L. (2000). Zebrafish kit mutation reveals primary and secondary regulation of melanocyte development during fin stripe regeneration. Development 127, 3715-3724. [DOI] [PubMed] [Google Scholar]
- Reddien P. W. and Sanchez Alvarado A. (2004). Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 20, 725-757. 10.1146/annurev.cellbio.20.010403.095114 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Oviedo N. J., Jennings J. R., Jenkin J. C. and Sanchez Alvarado A. (2005). SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327-1330. 10.1126/science.1116110 [DOI] [PubMed] [Google Scholar]
- Roberts D. S. and Linthicum F. H. Jr (2015). Distribution of melanocytes in the human cochlea. Otol. Neurotol. 36, e99-e100. 10.1097/MAO.0000000000000697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A. (2002). The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development 129, 5659-5665. 10.1242/dev.00167 [DOI] [PubMed] [Google Scholar]
- Scimone M. L., Srivastava M., Bell G. W. and Reddien P. W. (2011). A regulatory program for excretory system regeneration in planarians. Development 138, 4387-4398. 10.1242/dev.068098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone M. L., Kravarik K. M., Lapan S. W. and Reddien P. W. (2014). Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Rep. 3, 339-352. 10.1016/j.stemcr.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone M. L., Cote L. E., Rogers T. and Reddien P. W. (2016). Two FGFRL-Wnt circuits organize the planarian anteroposterior axis. Elife 5, e12845 10.7554/eLife.12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squarzoni P., Parveen F., Zanetti L., Ristoratore F. and Spagnuolo A. (2011). FGF/MAPK/Ets signaling renders pigment cell precursors competent to respond to Wnt signal by directly controlling Ci-Tcf transcription. Development 138, 1421-1432. 10.1242/dev.057323 [DOI] [PubMed] [Google Scholar]
- Stubenhaus B. M., Dustin J. P., Neverett E. R., Beaudry M. S., Nadeau L. E., Burk-McCoy E., He X., Pearson B. J. and Pellettieri J. (2016). Light-induced depigmentation in planarians models the pathophysiology of acute porphyrias. Elife 5, e14175 10.7554/eLife.14175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto H., Odani S. and Yamashita S. (1998). Cloning and expression of cDNA encoding rat liver 60-kDa lysophospholipase containing an asparaginase-like region and ankyrin repeat. J. Biol. Chem. 273, 12536-12542. 10.1074/jbc.273.20.12536 [DOI] [PubMed] [Google Scholar]
- Takeuchi S., Zhang W., Wakamatsu K., Ito S., Hearing V. J., Kraemer K. H. and Brash D. E. (2004). Melanin acts as a potent UVB photosensitizer to cause an atypical mode of cell death in murine skin. Proc. Natl. Acad. Sci. USA 101, 15076-15081. 10.1073/pnas.0403994101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A. J. and Erickson C. A. (2009). FOXD3 regulates the lineage switch between neural crest-derived glial cells and pigment cells by repressing MITF through a non-canonical mechanism. Development 136, 1849-1858. 10.1242/dev.031989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel J. C., Wagner D. E. and Reddien P. W. (2014). Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell 15, 326-339. 10.1016/j.stem.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Han X.-S., Li F.-F., Huang S., Qin Y.-W., Zhao X.-X. and Jing Q. (2016). Forkhead containing transcription factor Albino controls tetrapyrrole-based body pigmentation in planarian. Cell Discov. 2, 16029 10.1038/celldisc.2016.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witchley J. N., Mayer M., Wagner D. E., Owen J. H. and Reddien P. W. (2013). Muscle cells provide instructions for planarian regeneration. Cell Rep. 4, 633-641. 10.1016/j.celrep.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel O., Cote L. E., Poirier A., Satija R., Regev A. and Reddien P. W. (2015). A generic and cell-type-specific wound response precedes regeneration in planarians. Dev. Cell 35, 632-645. 10.1016/j.devcel.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.-T. and Johnson S. L. (2006). Small molecule-induced ablation and subsequent regeneration of larval zebrafish melanocytes. Development 133, 3563-3573. 10.1242/dev.02533 [DOI] [PubMed] [Google Scholar]
- Zhu S. J., Hallows S. E., Currie K. W., Xu C. and Pearson B. J. (2015). A mex3 homolog is required for differentiation during planarian stem cell lineage development. Elife 4, e07025 10.7554/eLife.07025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.