Fig. 2.
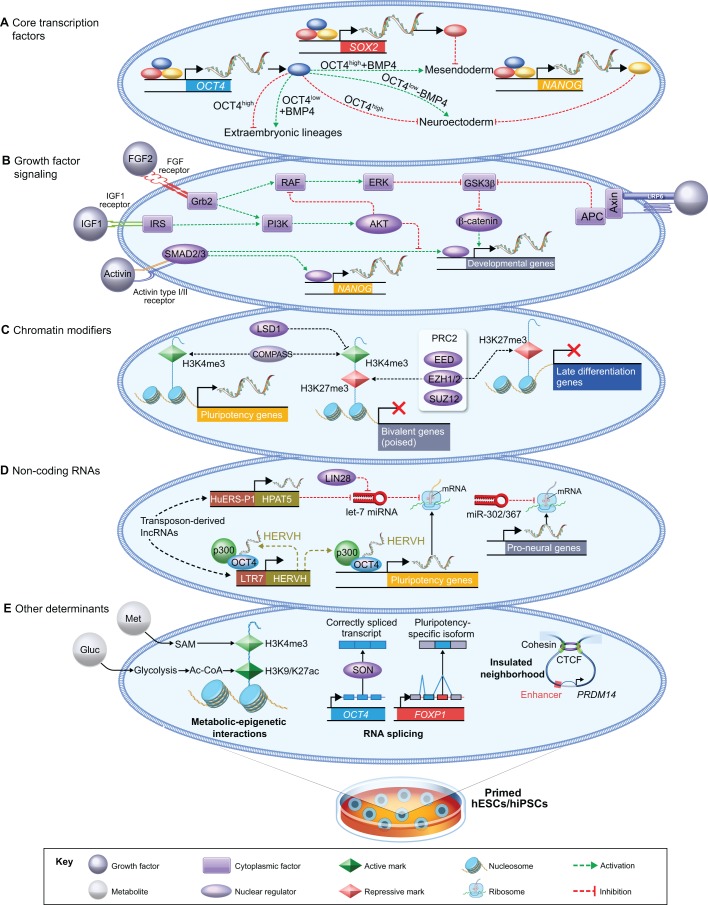
Layers of gene expression control in primed human PSCs. Overview of the major determinants of global gene expression in hPSCs cultured under conventional (primed) culture conditions. (A) The core transcription factors OCT4, SOX2 and NANOG form an autoregulatory network and repress distinct lineage fates in hESCs (Boyer et al., 2005; Wang et al., 2012). (B) Crosstalk among the major signaling pathways in hESCs as proposed by Singh et al. (2012). According to this model, activation of the PI3K/AKT pathway by IGF1 or FGF2 promotes the self-renewal of hESCs via two mechanisms. First, PI3K/AKT modulates the threshold of SMAD2/3 activity, allowing for the activation of NANOG but not mesendoderm-associated genes. As shown in A, the activation of NANOG stimulates the expression of core pluripotency genes and blocks neuroectoderm differentiation. Active PI3K/AKT also inhibits MEK/ERK and maintains GSK3β activity, which blocks β-catenin-mediated stimulation of pro-differentiation genes. Note that Wnt/β-catenin signaling has a distinct role in mESCs, where it functions to promote self-renewal instead of differentiation (ten Berge et al., 2011; Wray et al., 2011; Yi et al., 2011). (C) Chromatin-modifying enzymes with functional significance in hESC self-renewal include EZH1/2, which deposit H3K27me3 (Collinson et al., 2016; Shan et al., 2017), and enzymes that control the levels of H3K4me3 (Adamo et al., 2011; Bertero et al., 2015). (D) Various non-coding RNAs regulate the human pluripotent state. Transposon-derived lncRNAs, including HERVH and HPAT5, contribute to the self-renewal of hESCs (Lu et al., 2014; Durruthy-Durruthy et al., 2016a), as does miR-302/367 (Rosa et al., 2009; Lipchina et al., 2011; Rosa and Brivanlou, 2011). However, let-7 miRNA blocks the processing of pluripotency transcripts and is inhibited by LIN28 (Viswanathan et al., 2008; Newman et al., 2008; Rybak et al., 2008). (E) Other major determinants of gene expression in hPSCs. (Left) Metabolites, such as methionine (Met) and glucose (Gluc), generate substrates for histone modifications in hPSCs (Shiraki et al., 2014; Moussaieff et al., 2015). (Middle) The spliceosome produces a pluripotency-specific isoform of the transcription factor FOXP1, while SON ensures the accurate splicing of OCT4 and PRDM14 (Gabut et al., 2011; Lu et al., 2013). (Right) Insulated neighborhoods established by cohesion-associated CTCF loops constrain enhancer-promoter interactions at human pluripotency loci (Ji et al., 2016).