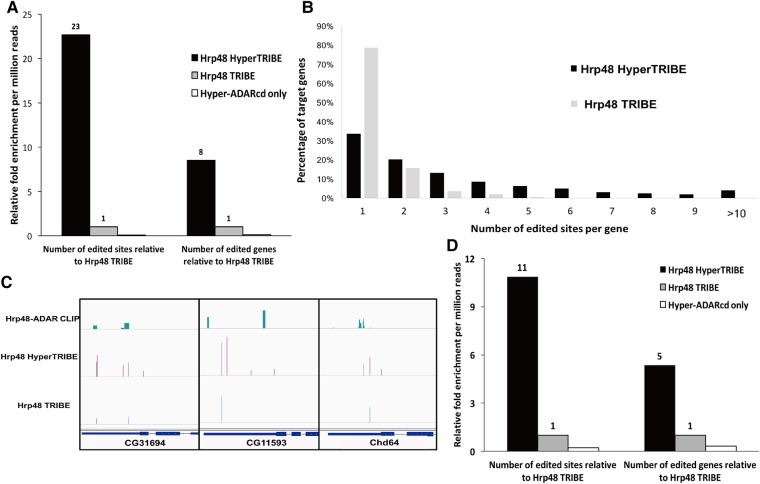
FIGURE 1.
HyperTRIBE dramatically increases both the number of target genes and the number of edited sites compared to TRIBE. (A) Although HyperTRIBE and TRIBE increases in both the number of editing events and genes edited in S2 cells, the increases are much more dramatic in HyperTRIBE-expressing cells. Numerically, HyperTRIBE identifies 10,689 common edited sites in two replicates, whereas the corresponding number for Hrp48 TRIBE is 291. Both TRIBE constructs have a reproducibility of around 60% and only replicable editing events are reported. There is no comparable increase in editing sites or genes with expression of the Hyper-ADARcd alone (11 sites identified). The number of genes and sites identified are normalized to the sequencing depth of each sample and are measured by relative fold change compared to TRIBE (see also Materials and Methods). (B) A much larger fraction of target genes are edited at multiple sites by HyperTRIBE than by TRIBE. The histogram indicates the percentage of target genes containing one to more than 10 editing sites. Genes with multiple sites may be transcripts bound more stably by Hrp48. (C) Three examples of commonly identified genes by HyperTRIBE, TRIBE, and CLIP are shown in the IGV genome browser. Hrp48-ADARcd CLIP data are from a previous publication (McMahon et al. 2016). Typically, the multiple editing sites in the HyperTRIBE cluster near the original sites identified by TRIBE. The height of the bars indicates editing frequency for TRIBE data and CLIP signal strength for CLIP data. (D) Expression of HyperTRIBE in all neurons (with the elav-gsg-Gal4 driver) in fly brains also increases the number of editing sites and genes compared to TRIBE. Editing events are identified and normalized as described in A.