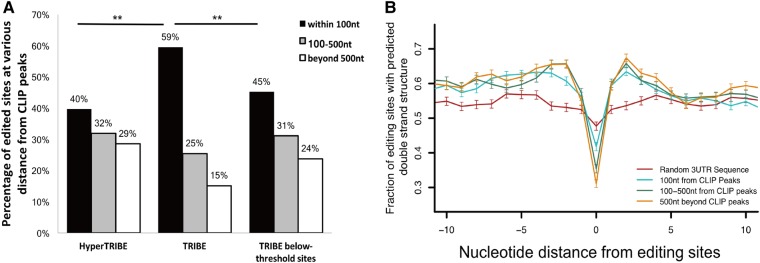
FIGURE 5.
HyperTRIBE edits distant adenosines more efficiently than TRIBE. (A) Bar graph showing the distribution of editing sites relative to Hrp48-ADARcd CLIP peaks. Editing sites are categorized into three classes according to their distance from Hrp48-ADARcd CLIP peaks: within 100 nt of CLIP peaks, between 100–500 nt and greater than 500 nt (McMahon et al. 2016). A smaller fraction of editing sites in HyperTRIBE is within 100 nt of CLIP peaks (40% versus 59%). (Z-test performed, [**] P < 0.0001, HyperTRIBE compared with TRIBE.) The below-threshold TRIBE sites show a similar pattern as HyperTRIBE sites and are more distant than above-threshold TRIBE sites. (Z-test performed, [**] P < 0.0001, TRIBE editing sites compared to TRIBE below-threshold sites.) (B) Editing sites that are closer to Hrp48 binding sites (CLIP peaks) have less local structure requirements compared to more distant sites. HyperTRIBE editing sites are categorized by their distance to Hrp48-ADARcd CLIP peaks as described in A. Sites within 100 nt of CLIP peaks (turquoise) show less flanking double-stranded structure and a less pronounced bulge at the edited adenosines than the 100–500 nt group (green) and beyond 500 nt group (orange). However, all three groups exhibit more surrounding double-stranded structures and a more pronounced bulged adenosine than a random 3′ UTR sequence, indicating the presence of a local structural preference even for the close editing sites. One thousand editing sites were randomly selected from each group as the input for folding analysis (see Materials and Methods).