FIGURE 2.
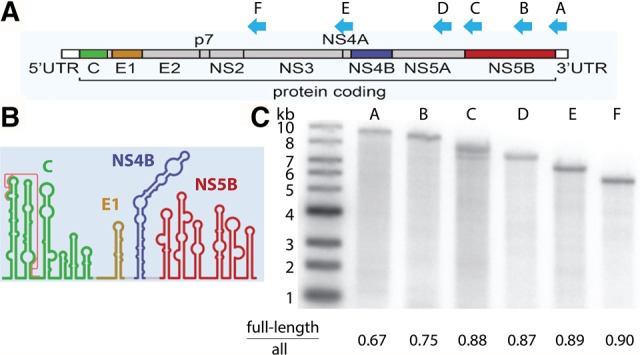
The RT reaction catalyzed by MarathonRT on a 9.6 kb viral genome. (A) Schematic diagram of HCV genome construction and (B) its secondary structure. Positions of primer binding sites (A [9461], B [8953], C [8051], D [7097], E [5912], F [4940]) are shown as blue arrows. (C) Representative denaturing alkaline agarose electrophoresis gel showing products of MarathonRT from multi-turnover RT reactions using full-length HCV genome as template. The ratio of signal intensity from full-length product divided by total products for each primer is indicated under each gel lane. The ladder is a double-stranded 1 kb DNA ladder (NEB), the mobility of which may be affected by incomplete denaturation in the gel.
