Figure 3. Association of De Novo Variants with TD Is Confirmed in the TSAICG Cohort.
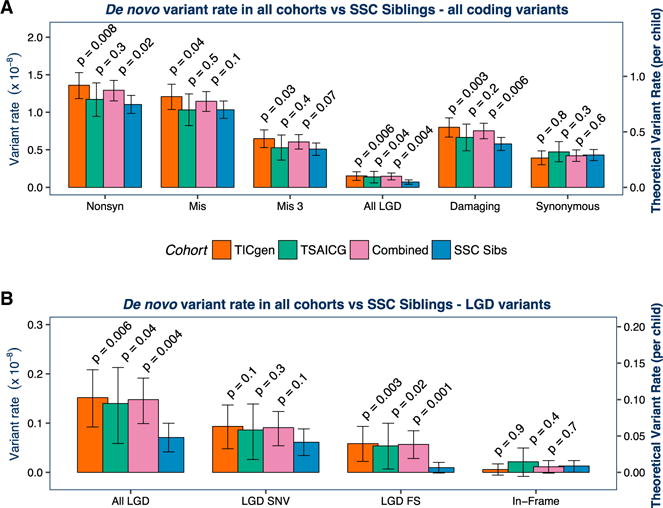
We next repeated the analyses in a non-overlapping cohort, ascertained and characterized by the TSAICG. De novo mutation rate per bp and theoretical mutation rate per child were calculated as in Figure 2. The TIC Genetics cohort is in red, TSAICG in green, the “Combined” TD cohort of TIC Genetics and TSAICG in purple, and the SSC control trios in blue. We compared the rate of de novo variants within the total callable exome with a one-sided rate ratio test (see Figure 2; Table 1). As in the TIC Genetics cohort, de novo LGD variants are elevated in TSAICG TD probands (p = 0.04) (A). De novo damaging variants as a group (LGD + Mis3) showed a trend toward enrichment in probands (p = 0.2). Again, FS indels occur at a substantially elevated rate (p = 0.02) (B). Neither synonymous de novo variants (p = 0.3; A) nor de novo in-frame indels (p = 0.4; B) showed any differences between TD and controls. Finally, we combined the TIC Genetics and TSAICG cohorts to obtain an overall estimate for de novo variant burden in TD (purple bars in A and B). De novo LGD variants are strongly associated with TD risk, occurring 2-fold more frequently in TD probands (RR 2.1, 95% CI 1.3–3.4, p = 0.004). De novo damaging variants (LGD + Mis3) are also associated (RR 1.3, 95% CI 1.1–1.5, p = 0.006). The distribution of de novo coding variants per individual in the TIC Genetics and TSAICG cohorts, as well as in the SSC siblings, follows an expected Poisson distribution (FigureS1). Mis3, missense variants predicted to be damaging by PolyPhen (Missense 3 or Mis3; PolyPhen2 [HDIV] score ≥ 0.957).
