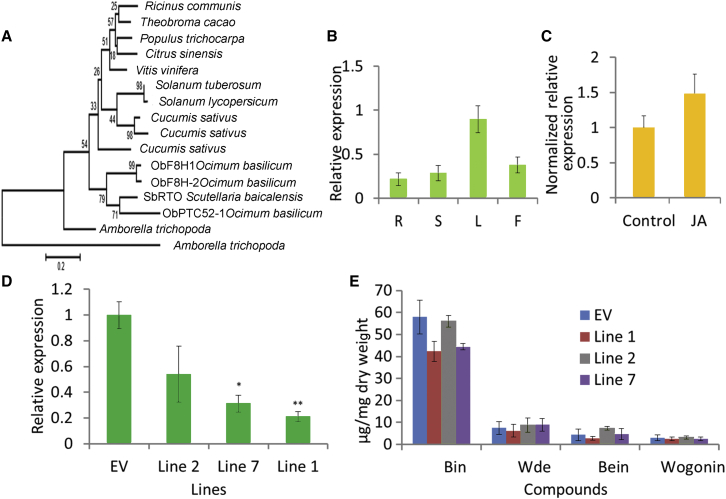
Figure 4.
Expression Patterns of SbRTO and the Effects of Silencing SbRTO by RNAi.
(A) Bootstrap consensus tree of RTO family. The maximum-likelihood method was used to construct this tree with 1000 replicate bootstrap support. Species names and GenBank IDs for peptide sequences used were: Ricinus communis, EEF2899; Theobroma cacao, EOY32295; Populus trichocarpa, EEE83915; Citrus sinensis, XP006488912; Vitis vinifera, XP002283592; Solanum tuberosum, XP006364436; Solanum lycopersicum, XP004237332; Cucumis sativus, XP004144089, XP004144087, XP004144144; Amborella trichopoda, ERN20453, ERN07127; Ocimum basilicum, AII16849 (ObF8H-1), AII16848 (ObF8H-2), AII16851(ObPTC52-1). For amino acid sequence alignments, see Supplemental Figure 10.
(B) Relative levels of SbRTO transcripts compared with β-actin determined by qRT–PCR analyses of cDNA from total RNA extracted from different S. baicalensis organs. R, roots; S, stems; L, leaves; F, flowers.
(C) Relative expression of SbRTO following MeJA treatment for 24 h. The expression levels were normalized to corresponding values from mock treatments.
(D) Silencing of SbRTO in different RNAi hairy root lines was measured by monitoring relative transcript levels by qRT–PCR.
(E) Measurements of RSFs from the SbRTO-RNAi lines used for transcript analyses. Bin, baicalin; Wde, wogonoside; Bein, baicalein; Win, wogonin.
SEs were calculated from three biological replicates. *P < 0.05 and **P < 0.01 (Student's t-test).