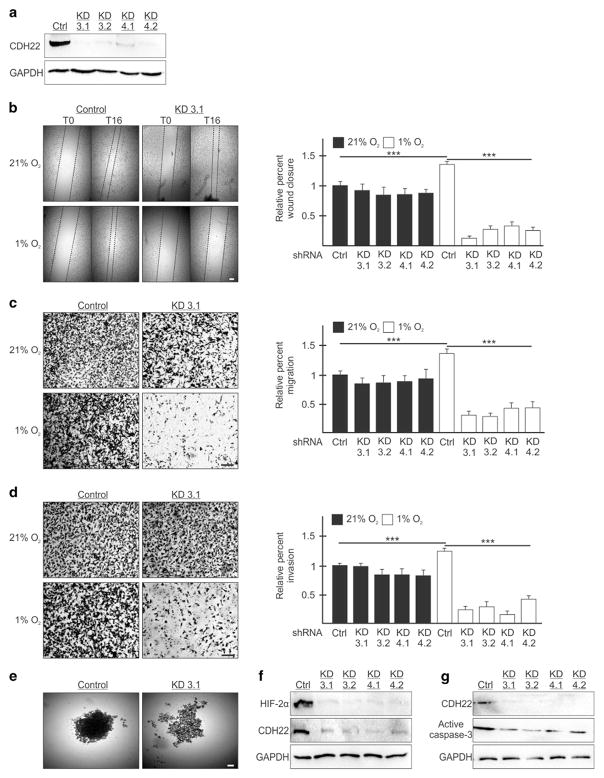
Figure 6.
Silencing CDH22 impairs hypoxic MDA-MB-231 cell migration, invasion and spheroid formation. (a) Western blot of CDH22 hypoxic protein levels in control (Ctrl) cells stably expressing a non-targeting shRNA or in cells stably expressing one of two shRNAs targeting CDH22 mRNA: Knockdown (KD) 3 and KD4. Two clones of each were generated: KD3.1, KD3.2, KD4.1 and KD4.2. GAPDH used as a loading control. (b) Control and CDH22 KD cells exposed to normoxia (21% O2) or hypoxia (1% O2) for 24 h followed by wound generation. Representative images at 0 h (T0) and 16 h (T16) after wound generation. (c, d) Transwell migration (c) and invasion (d) assays of control and CDH22 KD cells exposed to normoxia or hypoxia for 24 h. Representative images of transwell inserts 16 h after seeding and stained with crystal violet. (e) Light micrographs of spheroids composed of control cells or CDH22-depleted cells (KD3.1). (f, g) Western blot of hypoxia-inducible factor-2α (f) and active caspase-3 (g) protein levels in lysates of spheroids composed of control cells or CDH22-depleted cells. Data are presented relative to normoxic Ctrl as mean ±s.e.m., n ≥3, ***P<0.001, using a one-way ANOVA followed by Tukey’s HSD test. Scale bar, 100 μm.