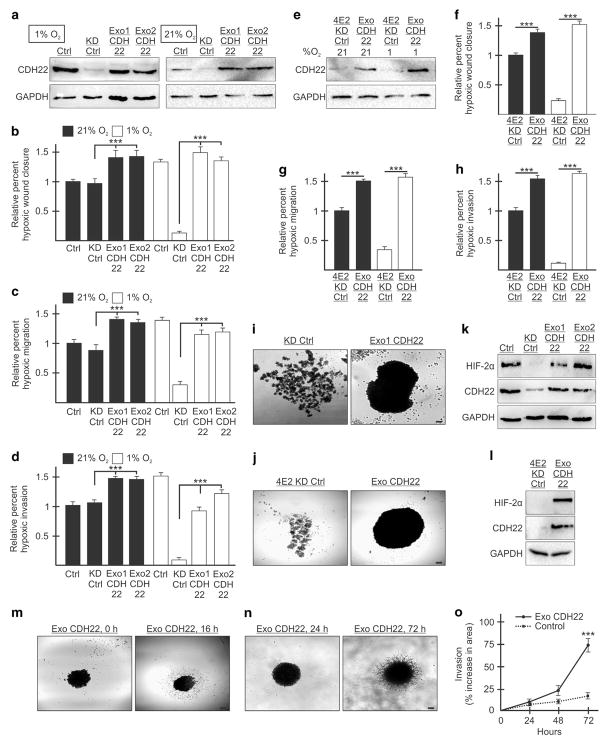
Figure 8.
Reintroduction of exogenous CDH22 restores the ability of MDA-MB-231 cells to migrate, invade and adhere to one another in hypoxia. (a) Western blot of control (Ctrl) cells stably expressing a non-targeting shRNA, CDH22 knockdown control (KD Ctrl) cells stably expressing shRNA targeting CDH22 mRNA and an exogenous empty vector or CDH22 codon-optimized coding sequence (clones Exo1 CDH22 and Exo2 CDH22) in hypoxia and normoxia. GAPDH used as a loading control. (b–d) Cells in (a) exposed to normoxia or hypoxia for 24 h followed by wound generation (b), or transwell migration (c) and invasion (d) assays. (e) Western blot of cells stably expressing shRNA targeting eIF4E2 and either an empty vector (4E2KD Ctrl) or exogenous CDH22 (Exo CDH22). (f–h) Cells in (e) exposed to normoxia or hypoxia for 24 h followed by wound generation (f), or transwell migration (g) and invasion (h) assays. (i, j) Spheroids composed of CDH22-depleted (i) or eIF4E2-depleted (j) control cells expressing empty vector or exogenous CDH22. (k, l) Western blot of hypoxia-inducible factor-2α protein in lysates of spheroids from (i) (k) and j (l). (m, n) Spheroid migration assay (m) and three-dimensional (3D) spheroid invasion assay (n) of spheroids composed of cells stably expressing exogenous CDH22. (o) Percent increase in spheroid area (invasion) in 3D invasion assay of spheroids stably expressing exogenous CDH22 compared with controls stably expressing non-targeting shRNA. Data are presented as mean ±s.e.m., n ≥3, ***P<0.001, using a one-way ANOVA followed by Tukey’s HSD test. Scale bar, 100 μm.