Abstract
Background
Influenza A virus (IAV) is the etiologic agent of the febrile respiratory illness, commonly referred to as ‘flu’. The lysosomal protease cathepsin B (CTSB) has shown to be involved in the lifecycle of various viruses. Here, we examined the role of CTSB in the IAV lifecycle.
Methods
CTSB-deficient (CTSB−/−) macrophages and the human lung epithelial cell line A549 cells treated with CA-074Me were infected with the A/Puerto Rico/8/34 strain of IAV (IAV-PR8). Viral entry and propagation were measured through quantitative real-time RT-PCR; production and localization of hemagglutinin (HA) protein in the infected host cells were analysed by Western blots, flow cytometry and confocal microscopy; production of progeny viruses were measured by a hemagglutination assay.
Results
CTSB−/− macrophages and CA-074Me-treated A549 cells had no defects in incorporating IAV-PR8 virions and permitting viral RNA synthesis. However, these cells produced significantly lower amounts of HA protein and progeny virions than wild-type or untreated cells.
Conclusion
These data indicate that CTSB is involved in the expression of IAV-PR8 HA protein and subsequent optimal production of IAV-PR8 progeny virions. Targeting CTSB can be a novel therapeutic strategy for treating IAV infection.
Keywords: Cathepsin B, Influenza A virus, CA-074 Me, Hemagglutinin
Introduction
Influenza virus is an enveloped virus with segmented, negative-sense, single-stranded RNA genome (1). Among the three species of influenza viruses A, B and C, human infections are most commonly caused by type A viruses (IAV), and result in a mild febrile illness to death (2). The genome of IAV encodes eleven proteins, eight of which are packaged into progeny virions, with hemagglutinin (HA) and neuraminidase (NA) expressed on the surface of virions as envelope glycoproteins. While NA facilitates progeny virion release, HA is responsible for virus entry (3–5) and other functions (6–8). Binding of the spike glycoprotein HA to sialic acid moieties on the host plasma membrane triggers the uptake of the virion into an endosome (4). The endosome is trafficked towards the nucleus and acidified as it matures (9). At ~5–6 pH, HA undergoes an irreversible conformational change to mediate the fusion of the viral envelope and endosomal membrane, thus releasing the ribonucleoprotein core into the host cytoplasm. Once in the cytoplasm, the core dissociates and the newly freed viral genome is actively imported into the nucleus, where replication of progeny genomes and transcription of mRNAs take place (10). Viral mRNAs are then exported from the nucleus for translation. Transmembrane proteins such as HA and NA are synthesized on rough endoplasmic reticulum, delivered to the plasma membrane via an exocytic pathway, and targeted to cholesterol-rich lipid raft microdomains where progeny virions are assembled and released (11). Accumulation of HA at the membrane triggers the nuclear export of progeny genomes to the membrane, mediated through the host cytoskeleton and Rab11-positive recycling endosomes (12–14). Although poorly understood, the process of budding and release is thought to begin with deformation of the host membrane, possibly due to HA-induced membrane curvature (15), interaction of the matrix M1 protein with progeny genomes (16, 17), and M2-mediated scission to separate a fully formed viral envelope (18). Recent studies also showed that IAV usurps host autophagy process to extracellularly release progeny viruses by directing autophagosomes to plasma membrane (19). Here, the proton channel matrix protein M2 plays a key role in directing autophagosomes to the plasma membrane, through directly interacting with the autophagy protein LC3 (20) or its proton pump activity (21). However, involvement of other autophagy processing factors in IAV exocytosis still remains to be examined.
Cathepsin B (CTSB) is a lysosomal cysteine protease primarily involved in the degradation of lysosomal proteins. In addition to its roles in protein turnover, CTSB plays key roles in the lifecycle of several viruses including Ebola virus (22), Nipah virus (23), Moloney murine leukemia virus (24) and feline coronavirus (25). CTSB catalytically activates viral membrane glycoproteins, which leads to viral release from endosomes to the cytoplasm through fusion of the viral envelope with the endosomal membrane (26, 27). Unenveloped reovirus uses CTSB for the proteolytic disassembly of the viral capsid while in host endosomes (28). Adeno-associated virus types 2 and 8 use CTSB to cleave capsid proteins, thereby priming rapid capsid disassembly in the nucleus (29). The catalytic activity of CTSB is also involved in the optimal replication of Herpes simplex virus type I DNA (30). We also showed that CTSB is involved in the trafficking of tumor necrosis factor α-containing vesicles to the plasma membrane (31) and efficient release of HIV virions from macrophages (32). Deficiency in functional CTSB retains tumor necrosis factor α and HIV virions in in the recycling endosomes and intracellular multivesicular bodies. Here, we further examined the role of CTSB in IAV infection using CTSB-deficient (CTSB−/−) bone marrow-derived immortalized macrophages (BMDIM) and CA-074Me in A549 human lung adenocarcinoma cells. We found that CTSB plays a key role in optimal production of progeny IAVs.
Methods
Reagents
The synthetic CTSB inhibitor [L-3-trans-(Propylcarbamoyl)oxirane-2-carbonyl]-L-isoleucyl-L-proline Methyl Ester (CA-074 Me) was purchased from Peptide Institute Inc. (Osaka, Japan). Actinomycin D (ActD; A9415) and LysoTracker Red (L-7528) were purchased from Sigma (Oakville, Canada) and Life Technologies (Invitrogen, Molecular Probes). The PR8 strain of IAV (A/Puerto Rico/8/34, H1N1) was grown in 10-day-old embryonated chicken eggs and used as infectious allantoic fluid. Primary antibodies used include polyclonal anti-β Actin (Rockland #600-401-886) and monoclonal anti-HA (hybridoma H28-E23) (33). Secondary antibodies used include goat anti-rabbit IgG IRDye 800 (Li-Cor #926-32211; Lincon, United States), goat anti-mouse IgG IRDye 800 (Li-Cor #926-32210; Lincon, United States), and fluorescein-conjugated goat anti-mouse IgG F (ab′)2 fragment (Jackson ImmunoResearch #115-096-146; West Grove, United States).
Cell culture
Human lung adenocarcinoma A549 cells were grown in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS; VWR; Mississauga, Canada), 1 mM MEM non-essential amino acid solution, 100 U/mL penicillin G, 100 μg/mL streptomycin and 1 mM sodium pyruvate (Sigma; Oakville, Canada). Primary and immortalized bone marrow-derived macrophages (BMDIM) from C57 BL/6 wild-type or CTSB−/− mice were prepared as described previously (31, 34). Cells were grown in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS; Sigma; Oakville, Canada), 1 mM MEM non-essential amino acid solution, 100 U/mL penicillin G, 100 μg/mL streptomycin and 1 mM sodium pyruvate. All cells were grown at 37 C in a humidified atmosphere containing 5% CO2.
Viral infections
One million cells were placed in 15 mL conical tubes and infected with IAV-PR8 at an MOI of 1, 5 or 10 for 1 h in 1 mL of PBS rotating at 37 C. Conditions with CA-074Me or ActD received the drug at the time of infection. After one hour adsorption, 5 mL of complete culture medium was added to the cells (with drug, if applicable) and incubation continued for a further 5 hours while rotating at 37 C.
MTT Assay
Seventy five thousand cells seeded in 96-well microtiter plates were treated in the presence or absence of drug for six hours at 37 C to reproduce conditions during infection. After treatment, MTT was added to a final concentration of 0.5 mg/mL and incubation continued for an additional 2 hours. Culture medium was then aspirated and 100 μL of DMSO was added to solubilize purple formazan crystals. After a 10 min incubation, absorbance was read at 570 nm using a plate reader, and the values of blank wells were averaged and subtracted from samples. Untreated conditions served as positive controls from which viability was calculated.
RT-qPCR
Briefly, RNA was extracted from cells using TriZol (Invitrogen; Burlington, Canada) and 0.5 μg was reverse transcribed with M-MuLV (New England Biolabs; Whitby, Canada) following manufacturer’s protocols using two viral gene-specific primers to yield template cDNA (see below). One μL of this sample was amplified with SYBR Green PCR Master Mix (Invitrogen; Burlington, Canada) and quantified using a Rotor-Gene RG3000 (Corbett Life Science; Kirkland, Canada) with the following cycling conditions: 94 C for 2 min; 94 C for 15 sec, 54 C for 30 sec, and 72 C for 30 sec with 30 cycles. Target amplicons for the two genes were PCR-amplified and a standard curve of each was generated by making a series of ten-fold dilutions. Known concentrations of amplicon were used to calculate copy number, and the resulting equations were used to determine the copy numbers of unknown samples based on Ct values as described previously (35, 36).
HA primers were generated from a sequence specific to the IAV-PR8 strain of influenza (GenBank: CY009447.1) using Primer3.
HA Fwd/RT primer: 5′-TGCTTCAAAACAGCCAAGTG-3′
HA Rev primer: 5′-GCCCAGTACCTGCTTCTCAG-3′
MA primers were generated from a sequence reported in literature to be highly conserved among viral subtypes (37).
Matrix Fwd/RT primer: 5′-CTTCTAACCGAGGTCGAAACG-3′
Matrix Rev primer: 5′-GCATTTTGGACAAAGCGTCT-3′
Western Blot
Samples were lysed on ice for 20 minutes in cold lysis buffer (50 mM tris-HCl [pH=7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS with protease inhibitor cocktail [Roche; Mississauga, Canada]) and run on 10% SDS gels at a voltage of 110V for 1.5 hours. Proteins separated by the gel were semi-dry transferred onto PVDF (Pall Life Sciences; Mississauga, Canada) for 1.5 hours at 18V. Membranes were blocked in 5% (w/v) skim milk in tris-buffered saline (TBS; 50 mM tris-HCl, 150 mM NaCl, pH 7.5) containing 0.08% Tween 20 (TTBS) for one hour and probed with primary antibodies at a 1:1000 dilution in TTBS overnight at room temperature. The following day, blots were washed in TTBS and incubated with secondary antibodies at a dilution of 1:10000 in TTBS for one hour. Blots were imaged using the Li-Cor Odyssey system (Guelph, Canada). Densitometric analysis on Western blots was performed using the densitometry feature in ImageJ (NIH; Bethesda, United States). Lanes were analyzed and the resulting histograms were used to measure band density by measuring the area under each peak with any background subtracted. Densities of target immunoreactivities were then normalized to a loading control (β-actin) for data analysis.
Flow cytometry analysis
BMDIM were treated with IAV-PR8 at an MOI of 1 with or without CA-074Me for 1h. After 6 h, cells were washed with ice-cold PBS, and then sequentially incubated with anti-HA antibodies and Alexa Fluor488-conjugated secondary antibody for 30 min each at 4 °C. Cells were then fixed with 1% par-formaldehyde and analyzed by flow cytometry using a FACS CantoII (BD Biosciences), Data analysis was performed using CellQuest software (BD Biosciences).
Immunofluorescence Microscopy
Fifty thousand A549 cells suspended in 50 μL of PBS were seeded onto coverslips and allowed to attach (~1 hour). Cells were then infected with PR8 at a high MOI (5 or ~10) by directly pipetting virus onto coverslips; any inhibitors indicated were added at this time. After a one hour adsorbtion period, 2 mL of warm media (with inhibitors, if applicable) was added to 6-wells containing coverslips and incubation was allowed to continue for an additional 4.5 hours. At this point, LysoTracker Red was added to a final concentration of 200 nM and cells were incubated for 30 min. After the six hour infection period was complete, cells were washed with PBS and fixed in 4% formaldehyde for 17 min. For conditions requiring permeabilization, cells were treated with 0.25% Triton X-100 for 10 min. After two washes in PBS, cells were blocked in PBS + 1% BSA + 0.05% Tween 20 for one hour and then incubated in anti-HA primary antibody at a 1:10 dilution overnight at 4 C. The following day, slides were washed in PBS and incubated in secondary (anti-mouse FITC) at a 1:100 dilution for two hours. Slides were then washed once more and incubated in Hoechst 33342 for 3 min at a 1:4000 dilution, rinsed, mounted, and viewed using the Zeiss LSM 510 confocal fluorescence microscope. Images were acquired using Zen 2008 (Zeiss; Toronto, Canada). Colocalization was assessed using the JACoP plugin for ImageJ (38).
Hemagglutination Assay
Hemagglutination assays were performed as described previously (39). Briefly, 1 x 106 BMDIM or A549 cells in the presence or absence of CA-074Me were infected at an MOI of 1 in a total volume of 1 mL PBS. After one hour adsorption, PBS and virus were aspirated, and cells were washed twice in PBS. Two mL of fresh serum-free DMEM with 2.5 μg/mL trypsin was added to A549 cells, along with CA-074Me if applicable, and incubation continued for an additional 23 h. Serum was omitted as it contains nonspecific inhibitors of IAV infection (40). Since BMDIM showed significant sensitivity to prolonged incubation in serum-free media, 2 mL of complete RPMI was added for 17 h, after which time media was removed and cells were washed twice in PBS. Two mL of fresh serum-free RPMI with 2.5 μg/mL trypsin was added for the remaining 6 h of the 23 h incubation. Twenty four hours post-infection, supernatant was collected and centrifuged to pellet debris. Fifty μL from each condition was added to 50 μL of PBS in triplicate and serially diluted two-fold. A PBS-only control and a positive control using the stock virus were run with each replicate. Fifty μL of a 0.5% adult chicken erythrocyte (Charles River Laboratories; Wilmington, United States) solution in PBS was added to each well and gently mixed. After incubating 30 min at room temperature, wells were scored as either positive or negative, and the titre reported as the reciprocal of the highest dilution which yielded agglutination.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 4 (GraphPad Software; San Diego, United States). Results were considered statistically significant if p ≤ 0.05 for either t-test or one-way ANOVA followed by a Tukey’s post hoc test, as stated in the figure legends. Data are presented as means ± SEM, where error bars denote biological variations between experiments. Co-localization was assessed using the JACoP plugin for ImageJ to determine Pearson’s coefficients to numerically define the degree of overlap between red and green channels.
RESULTS
CTSB B is required for optimal production of IAV-PR8
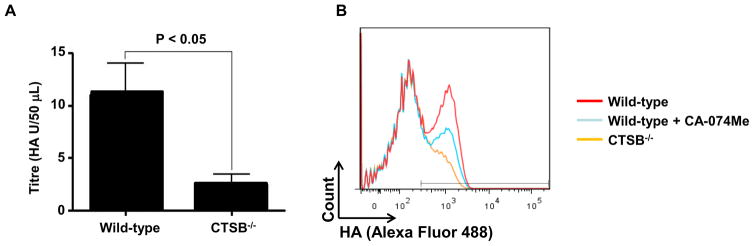
To examine the involvement of CTSB in IAV-PR8 propagation, we first examined production of progeny IAV virions by wild-type and CTSB-deficient (CTSB−/−) bone marrow-derived immortalized macrophages (BMDIM) through hemagglutination assays after 24 h post-infection. The release of progeny virions by CTSB−/− cells was about 4-fold less than that by wild-type cells (Fig. 1A).
Figure 1. Cathepsin B deficient (CTSB−/−) leads to defective production of progeny IAV-PR8 virions and HA presentation.
(A) Wild-type and CTSB−/− BMDIM (1.0 x106) were infected with IAV-PR8 (MOI of 1) for 24 h. Cell culture supernatant of the last 6 h of infection were collected and subjected to a standard hemagglutination assay using chicken red blood cells and a series of two-fold dilutions of the supernatants. After 30 min incubation, wells were scored as being either positive or negative for agglutination, and the titer expressed as the reciprocal of the highest dilution yielding agglutination. The titre of CTSB−/− BMDIM was significantly less than that of wild-type cells (p > 0.05; n=4). (B) Wild-type and CTSB−/− BMDIM (1.0 x106) were infected with IAV-PR8 (MOI of 1) with or without CA-074Me (50 μM) for 1 h as described in Methods. After 6 h of infection, surface expression of HA protein was analyzed using flow cytometry. Data shown are representative results of similar observations in more than two independent experiments.
CTSB-deficient or CA-074Me-treated BMDIM are defective in presenting HA on the plasma membrane
Since CTSB−/− cells were defective in releasing IAV-PR8 virions, we examined whether CTSB was involved in presentation of HA on the surface of infected cells. To this end, wild-type cells, wild-type cells treated with CA-074Me (50 μM, for 30 min) and CTSB−/− cells were infected with IAV-PR8 at an MOI of 2 for 5 hours, and amounts of HA on the cell surface were analyzed using FACS. In wild-type non-treated cells, about 50% of cells were labeled with HA antibodies (Fig. 1B, red line). However, the numbers of HA-labeled cells were decreased to about 28% and 10% in CA-074Me treated (green line) and CTSB−/− cells (orange line), respectively. These results suggest that CTSB or its catalytic activity was required for proper plasma membrane presentation of IAV-PR8 HA proteins.
CTSB is required for optimal production of HA protein
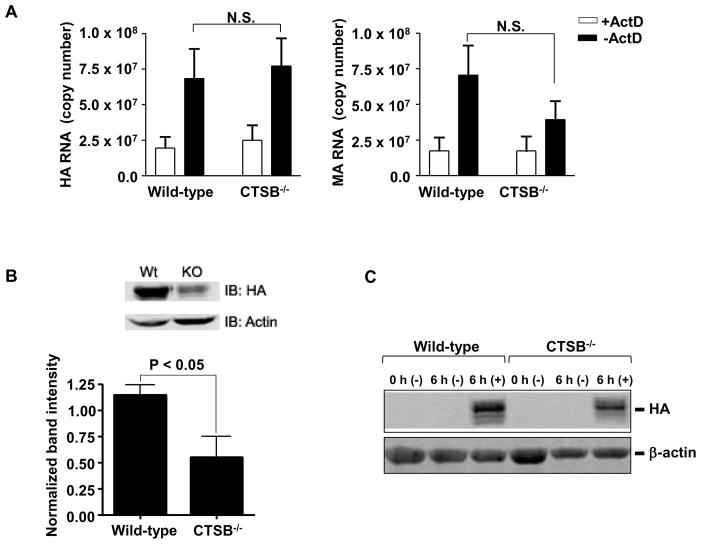
To further examine whether CTSB was involved in the entry or replication of IAV-PR8, wild-type and CTSB−/− cells were infected in the presence or absence of actinomycin D (ActD). ActD is a transcriptional inhibitor that has been used extensively in influenza research to block viral replication by preventing nuclear import of the viral genome (41, 42). Therefore, cells treated with ActD contain only the genetic material and viral proteins endocytosed from the original inoculum, affording an accurate means of assessing viral endocytosis in the absence of replication products. The extents of viral entry and replication were determined by real-time RT-qPCR on two viral genes: the hemagglutinin (HA) specific to the PR8 strain of IAV and the matrix (MA) gene highly conserved among IAV subtypes (37). RT-qPCR has proven to be a useful tool in quantifying influenza particles (43, 44). This is due in part to its sensitivity and the genetic nature of the virus, which packages one copy of each viral gene per virion. Thus, calculations of copy number accurately reflect the absolute number of virions present. As shown in Fig. 2A, RNA copy numbers of HA (left panel) and MA (right panel) were similar between wild-type and CTSB−/− cells treated with ActD. Both copy numbers of HA and MA were ~2-fold higher in cells without ActD treatments, indicating production of new viral RNAs. ActD did not influence the viability of these cells (Data not shown). Since CTSB was not involved in either viral entry or RNA synthesis. We examined whether HA protein expression was defected in CTSB−/− cells. To this end, cell lysates of IAV-infected wild-type and CTSB−/− cells in the presence or absence of ActD were subjected to Western blot analysis for viral HA protein. Densitometric analysis of the immunereacted bands was performed and the intensities were normalized to those of β-actin. As shown in Fig. 2B, the amount of HA protein detected was significantly lower (~50%) in CTSB−/− cells than that of wild-type cells. Similar results were also obtained in primary bone marrow-derived macrophages prepared from wild-type and CTSB−/− mice (Fig. 2C).
Figure 2. CTSB−/− BMDIM are normal in up taking IAV-PR8 virions and propagating viral RNAs, but defected in expression of HA protein.
A. Wild-type and CTSB−/− BMDIM were infected with influenza A PR8 for 6 h at an MOI of 1 in the presence or absence of actinomycin D (ActD; 5 μg/mL). Cells were then harvested and total RNA extracts were used for RT-qPCR analysis for hemagglutinin (HA; PR8-specific) and matrix protein (MA; pan-specific). Data are expressed as means ± SEM from at least three independent experiments (N.S. = not significant). B–C. Similarly, BMDIM (B) and primary bone marrow-derived macrophages (C) were infected, harvested, lysed and subjected to Western blotting for the influenza HA protein. B. A representative Western blot for HA protein and β-actin (as a loading control) is presented (top panel). Densitometric analysis of the bads was performed and results are expressed as means ± SEM (n = 3; bottom panel). C. A representative Western blot for HA protein and β-actin from two separate experiments is presented.
CA-074Me inhibits IAV-PR8 HA production in human lung epithelial cell line A549 cells
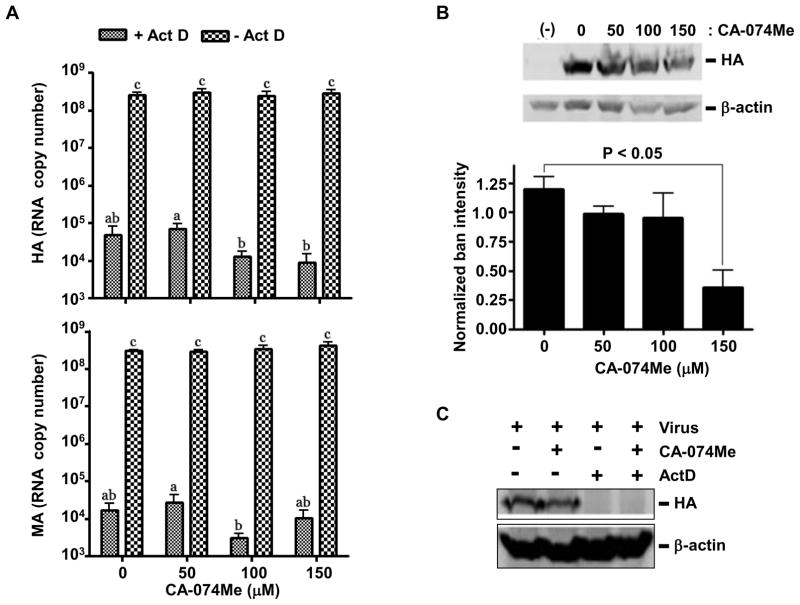
To examine whether the involvement of CTSB in IAV-PR8 production was specific to murine macrophages or not, we performed similar experiments using CA-074Me in the human lung epithelial cell line A549 cells. These cells more closely approximate the respiratory epithelium which is infected in vivo, and as such are commonly used in influenza research. Consistent with data obtained from CTSB−/− BMDIM, CA-074Me (up to 150 μM) had no effects on viral RNA copy numbers of HA and MA in IAV-PR8-infected A549 cells in the absence or presence of ActD (Fig. 3A). However, Western blots against HA protein showed significantly decreased band intensities (~25%) in cells treated with 150 μM of CA-074Me, compared to those in non-treated cells (Fig. 3B). Since no HA bands were detected in cells treated with ActD, HA proteins shown in Western blots likely represent newly synthesized HA proteins (Fig. 3C). Treatments of CA-074Me had no effects on the viability of A549 cells (Data not shown).
Figure 3. CA-074Me inhibits expression of HA protein without affecting IAV-PR8 virion uptake or viral RNA propagation in A549 cells.
A. A549 cells pretreated with different concentrations of CA-074Me for 30 min were infected with influenza A virus strain PR8 for 6 h at an MOI of 1 in the presence or absence of actinomycin D (ActD; 5 μg/mL). Cells were then harvested and total RNA extracts were used for RT-qPCR analysis against hemagglutinin (upper panel) and matrix (lower panel) RNAs. Data are expressed as means ± SEM from at least three independent experiments. Columns accompanied by the same letter are not significantly different from each other (Tukey’s post hoc test, p < 0.05). B. Similarly, A549 cells treated with CA-074Me at doses indicated were infected, harvested, lysed and subjected to Western blotting for the influenza HA protein, and the resulting blots were used for densitometric analysis. A representative Western blot for HA protein and β-actin (as loading control) is presented (upper panel). Data are expressed as means ± SEM from at least three independent experiments (lower panel). C. A Western blot for HA protein in A549 cells treated with CA-074-Me (150 μM) or ActD (5 μg/mL) is presented.
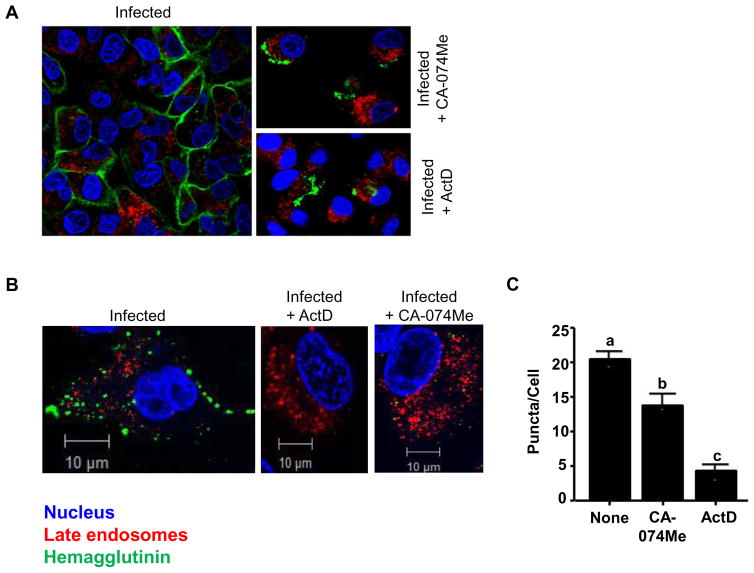
CA-074Me prevents formation of HA positive puncta in IAV-PR8-infected A549 cells
After translation of HA RNAs, HA proteins are trafficked to the host plasma membrane via the Golgi network where they are embedded on the cell surface (45). Since CTSB has been shown to have a role in trafficking functions (31, 32, 46), it is possible that its inhibition may disrupt the transport of HA, subsequently targeting cargo for destruction in lysosomes (47). Thus, we examined whether HA proteins were prevented from being transported to plasma membrane or redirected to lysosomal degradation, which may result in a defect in the infection of adjacent cells and an overall decrease in HA protein production by CA-074Me. To this end, A549 cells were infected in the presence or absence of CA-074Me and/or ActD, and HA proteins were visualized through confocal immunofluorescence microscopy. In non-treated cells, IAV-PR8 infection (MOI of 10) yielded an even staining pattern for HA (green) in the plasma membrane of many cells (Fig. 4A, left panel). Treatment with either CA-074Me (150 μM) or ActD (5 μg/mL) abolished the staining pattern, and instead only aggregated puncta at the surface of cells were visible in a limited number of cells (right panels). Since no HA was detected in cells treated with ActD through Western blots (Fig. 3C), the aggregated puncta detected in CA-074Me- or ActD-treated cells could be viruses inoculated. To eliminate signals from inoculated viruses and to quantify production of HA-positive vesicles, A549 cells were infected with IAV-PR8 at an MOI of 1 and cells were permeabilized before immunostaining. As shown in Fig. 4B, non-treated cells showed distinct punta, whereas CA-074Me- or ActD-treated cells showed no aggregated puncta and significantly less intracellular staining for HA relative to untreated cells; ActD-treated cells showed almost no intracellular staining (Fig. 4B, middle and right panels; Fig. 4C). Furthermore, colocalization of HA and late endosome/lysosome was quantified using the analysis described in (48) and found that there was no apparent colocalization of these compartments in any of the conditions (Pearson’s coefficients = 0.057, 0.082 and 0.003 for untreated, CA-074Me-treated and ActD-treated cells, respectively).
Figure 4. CA-074Me inhibits surface expression of HA protein in A549 cells.
A. A549 cells adhered to coverslips were infected for six hours at an MOI of 10 in the presence of PBS (left), CA-074Me (150 μM; top right), ActD (5 μg/mL; bottom right). About 6 h post-infection, cellular compartments were labelled for late endosomes/lysosome (Lysotracker Red®), HA (green) and nucleus (Hoechst 33342; blue), as described in Methods. Cells were then viewed using a Zeiss LSM 510 confocal fluorescence microscope and images shown are representative cells from respective treatments. B. Similarly, cells were processed as in A, except that cells were infected with IAV-PR8 at MOI of 5 and permeabilized with 0.25% Triton X-100 before HA labelling. Images shown are representative images from three independent experiments. C. The numbers of puncta per cell in random fields of view were quantified and plotted for each treatment group. Data are expressed as means ± SEM (p > 0.05; one-way ANOVA; n=3). Columns accompanied by the same letter are not significantly different from each other by Tukey’s post hoc test.
Discussion
Ours and several other studies have identified CTSB as a key factor involved in the progress of various viral lifecycles (22–26, 29, 30, 32, 49). In most cases, the role of CTSB is to proteolytically activate viral proteins which facilitate escape into the cytoplasm following endocytosis. Given that IAV uses endosomes to enter host cells and transport viral genome to nucleus, we examined the role of CTSB in the early stages of the viral lifecycle. However, no significant differences between wild-type and CTSB−/− BMDIMs, and between CA-074Me-treated and non-treated A549 cells were detected in viral entry and subsequent replication (Fig. 2B & 2A). These data indicate that CTSB played no apparent role in the early lifecycle of IAV-PR8. As expected, cells treated with ActD had lower copy numbers of both HA and MA RNAs than untreated in BMDIM and A549 cells. However, there was a notable difference in the titres achieved in these cells in terms of both entry and replication. In the presence of ActD, A549 cells show fewer copies of both genes (~1–5 x 104) compared to BMDIM (~2 x 107). At an MOI of 1, the viral inoculum contained approximately 106 particles, suggesting that approximately 5% of inoculum had infected A549 cells, while BMDIM contained 20-fold more viral RNA copies than inoculated. The large amount of viral RNAs in BMDIM could be due to the phagocytic nature of the cell and the propensity of influenza virus to generate defective particles in culture (45, 50, 51). Conversely, in the absence of ActD, A549 cells produced about 5-fold more viral RNAs than BMDIM (~7 x 107 versus ~3 x 108). This may reflect the permissiveness of the respective cell type to infection, which is consistent with a previous observation that IAV replicates to higher titres in epithelial cells than macrophages (52).
Out of the eleven proteins generated during IAV infection, HA is absolutely crucial for infectivity (5) and virions lacking HA are non-infectious due to an inability to bind host cells or escape endosomes following endocytosis (53). HA proteins are also abundantly expressed in infected cells (45) and easily detected by Western blot or cytometry analysis. Therefore, HA expression was examined as a marker for viral protein production. Approximately 2.5 h post-infection, the background of host proteins is dramatically reduced as the cellular machinery is usurped for the production of viral proteins. Most viral proteins including HA are detected in 3 h post infection, which is stable up to 10 h (54). Unlike the case for the entry and replication, CTSB−/− BMDIM or CA-074Me-treated A549 cells contained significantly less amounts of HA protein than wild-type or non-treated cells, respectively, when measured 6 h post-infection (Fig. 2B–C & 3B). Also, HA proteins were not detected in infected cells in the presence of ActD (Fig. 3C), suggesting that immunoreactive HA bands detected newly synthesized viral proteins. Collectively, these data suggest that CTSB was required for optimal production of IAV-PR8 HA protein.
To examine the fate of HA protein in CTSB-deficient cells, A549 cells infected with IAV-PR8 in the presence or absence of CA-074Me or ActD were visualized using confocal immunofluorescence microscopy. We found that CA-074Me-treated cells had a noticeable reduction in the number and size of HA-positive puncta which were not colocalized with late endosomes/lysosomes (Fig. 4), suggesting that HA proteins were not likely targeted to lysosomal degradation in cells defected in CTSB activity. The lysosomal degradation pathway inhibitor chloroquine also did not induce an accumulation of HA proteins in cells treated with CA-074Me-treated or in CTSB−/− cells (data not shown); however, any inhibition in endosomal maturation could inhibit viral release from endosomes to the host cytoplasm (55), obscuring the data interpretation. The ubiquitin-proteasome degradation pathway is an alternative process for protein degradation; however, the proteosome inhibitor MG132 also did not cause an increase of HA protein expression in IAV-PR8-infected CTSB−/− BMDIM cells (data not shown). Therefore, further studies are required to determine how CTSB enhances production of HA proteins at the levels of protein degradation or synthesis.
Nonetheless, it is clear that CTSB was required for optimal production of HA proteins, and genetic defects in or chemical inhibition of CTSB expression resulted in impairments of the surface presentation of HA protein and progeny virion production (Fig. 1 ). To measure progeny virus production we used the hemagglutination assay which measures viral titres on the basis of HA protein on the surface of particles. Progeny virions acquire HA and other envelope glycoproteins passively by budding from infected host cells which bear these proteins on their membranes (56). Absence of HA in the host membrane could have several deleterious consequences, considering HA is critical for endocytosis (5) and endosomal escape (57), and export and packaging of progeny genomes (7, 8). Thus, if progeny lacking HA are released from infected cells, it is likely that they will possess defects related to one or more of the above functions.
Resistance of circulating IAV strains to older antivirals such as amantadine as well as newer antivirals such as oseltamivir has increased at an alarming rate over the past decade (58). Given the rate with which antiviral therapies are becoming obsolete due to viral mutations, a more effective strategy would be to target host rather than viral components required for the effective replication of IAV. Previously, IAV was shown to elevate CTSB activity and expression that facilitates antigen processing in dendritic cells murine cells (59). Here, we also showed that deficiency in CTSB lead to defects in IAV virion production, suggesting that targeting CTSB could be a novel therapeutic strategy for IAV infection.
Acknowledgments
This work was supported by Canadian Institute of Health Research Operating Grant MOP93551 to S.O.K, HBF134179 to SDB & MOP130465 to SMMH.
Footnotes
Authors’ contributions
MDC, SOK – conceived & designed the experiments, analyzed data, and wrote the paper. MDC, SDH – performed the experiments. SMMH & SDB– provided IAV and reagents, and participated in designing experiments. All authors edited the manuscript and approved the final manuscript.
References
- 1.Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zambon MC. Epidemiology and pathogenesis of influenza. J Antimicrob Chemother. 1999;44(Suppl B):3–9. doi: 10.1093/jac/44.suppl_2.3. [DOI] [PubMed] [Google Scholar]
- 3.Liu C, Eichelberger MC, Compans RW, Air GM. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J Virol. 1995;69:1099–1106. doi: 10.1128/jvi.69.2.1099-1106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rust MJ, Lakadamyali M, Zhang F, Zhuang X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat Struct Mol Biol. 2004;11:567–573. doi: 10.1038/nsmb769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang S, Li R, Du L, Liu S. Roles of the hemagglutinin of influenza A virus in viral entry and development of antiviral therapeutics and vaccines. Protein Cell. 2010;1:342–354. doi: 10.1007/s13238-010-0054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Leser GP, Pekosz A, Lamb RA. The cytoplasmic tails of the influenza virus spike glycoproteins are required for normal genome packaging. Virology. 2000;269:325–334. doi: 10.1006/viro.2000.0228. [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Pekosz A, Lamb RA. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J Virol. 2000;74:4634–4644. doi: 10.1128/jvi.74.10.4634-4644.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marjuki H, Alam MI, Ehrhardt C, Wagner R, Planz O, Klenk HD, Ludwig S, Pleschka S. Membrane accumulation of influenza A virus hemagglutinin triggers nuclear export of the viral genome via protein kinase Calpha-mediated activation of ERK signaling. J Biol Chem. 2006;281:16707–16715. doi: 10.1074/jbc.M510233200. [DOI] [PubMed] [Google Scholar]
- 9.Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. Visualizing infection of individual influenza viruses. Proc Natl Acad Sci U S A. 2003;100:9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helenius A. Unpacking the incoming influenza virus. Cell. 1992;69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 11.Leser GP, Lamb RA. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology. 2005;342:215–227. doi: 10.1016/j.virol.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 12.Eisfeld AJ, Kawakami E, Watanabe T, Neumann G, Kawaoka Y. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J Virol. 2011;85:6117–6126. doi: 10.1128/JVI.00378-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jo S, Kawaguchi A, Takizawa N, Morikawa Y, Momose F, Nagata K. Involvement of vesicular trafficking system in membrane targeting of the progeny influenza virus genome. Microbes Infect. 2010;12:1079–1084. doi: 10.1016/j.micinf.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 14.Momose F, Sekimoto T, Ohkura T, Jo S, Kawaguchi A, Nagata K, Morikawa Y. Apical transport of influenza A virus ribonucleoprotein requires Rab11-positive recycling endosome. PLoS One. 2011;6:e21123. doi: 10.1371/journal.pone.0021123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen BJ, Leser GP, Morita E, Lamb RA. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J Virol. 2007;81:7111–7123. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ali A, Avalos RT, Ponimaskin E, Nayak DP. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J Virol. 2000;74:8709–8719. doi: 10.1128/jvi.74.18.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noton SL, Medcalf E, Fisher D, Mullin AE, Elton D, Digard P. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J Gen Virol. 2007;88:2280–2290. doi: 10.1099/vir.0.82809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munz C. The Autophagic Machinery in Viral Exocytosis. Front Microbiol. 2017;8:269. doi: 10.3389/fmicb.2017.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beale R, Wise H, Stuart A, Ravenhill BJ, Digard P, Randow F. A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe. 2014;15:239–247. doi: 10.1016/j.chom.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren Y, Li C, Feng L, Pan W, Li L, Wang Q, Li J, Li N, Han L, Zheng X, Niu X, Sun C, Chen L. Proton Channel Activity of Influenza A Virus Matrix Protein 2 Contributes to Autophagy Arrest. J Virol. 2015;90:591–598. doi: 10.1128/JVI.00576-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diederich S, Thiel L, Maisner A. Role of endocytosis and cathepsin-mediated activation in Nipah virus entry. Virology. 2008;375:391–400. doi: 10.1016/j.virol.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar P, Nachagari D, Fields C, Franks J, Albritton LM. Host cell cathepsins potentiate Moloney murine leukemia virus infection. J Virol. 2007;81:10506–10514. doi: 10.1128/JVI.02853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regan AD, Shraybman R, Cohen RD, Whittaker GR. Differential role for low pH and cathepsin-mediated cleavage of the viral spike protein during entry of serotype II feline coronaviruses. Vet Microbiol. 2008;132:235–248. doi: 10.1016/j.vetmic.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gnirss K, Kuhl A, Karsten C, Glowacka I, Bertram S, Kaup F, Hofmann H, Pohlmann S. Cathepsins B and L activate Ebola but not Marburg virus glycoproteins for efficient entry into cell lines and macrophages independent of TMPRSS2 expression. Virology. 2012;424:3–10. doi: 10.1016/j.virol.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P, Kawaoka Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebert DH, Deussing J, Peters C, Dermody TS. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J Biol Chem. 2002;277:24609–24617. doi: 10.1074/jbc.M201107200. [DOI] [PubMed] [Google Scholar]
- 29.Akache B, Grimm D, Shen X, Fuess S, Yant SR, Glazer DS, Park J, Kay MA. A two-hybrid screen identifies cathepsins B and L as uncoating factors for adeno-associated virus 2 and 8. Mol Ther. 2007;15:330–339. doi: 10.1038/sj.mt.6300053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Link MA, Silva LA, Schaffer PA. Cathepsin B mediates cleavage of herpes simplex virus type 1 origin binding protein (OBP) to yield OBPC-1, and cleavage is dependent upon viral DNA replication. J Virol. 2007;81:9175–9182. doi: 10.1128/JVI.00676-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ha SD, Martins A, Khazaie K, Han J, Chan BM, Kim SO. Cathepsin B is involved in the trafficking of TNF-alpha-containing vesicles to the plasma membrane in macrophages. J Immunol. 2008;181:690–697. doi: 10.4049/jimmunol.181.1.690. [DOI] [PubMed] [Google Scholar]
- 32.Ha SD, Park S, Hattlmann CJ, Barr SD, Kim SO. Inhibition or deficiency of cathepsin B leads defects in HIV-1 Gag pseudoparticle release in macrophages and HEK293T cells. Antiviral Res. 2012;93:175–184. doi: 10.1016/j.antiviral.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 33.Gerhard W, Yewdell J, Frankel ME, Webster R. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature. 1981;290:713–717. doi: 10.1038/290713a0. [DOI] [PubMed] [Google Scholar]
- 34.Adami C, Brunda MJ, Palleroni AV. In vivo immortalization of murine peritoneal macrophages: a new rapid and efficient method for obtaining macrophage cell lines. J Leukoc Biol. 1993;53:475–478. doi: 10.1002/jlb.53.4.475. [DOI] [PubMed] [Google Scholar]
- 35.Shelton H, Smith M, Hartgroves L, Stilwell P, Roberts K, Johnson B, Barclay W. An influenza reassortant with polymerase of pH1N1 and NS gene of H3N2 influenza A virus is attenuated in vivo. J Gen Virol. 2012;93:998–1006. doi: 10.1099/vir.0.039701-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ngaosuwankul N, Noisumdaeng P, Komolsiri P, Pooruk P, Chokephaibulkit K, Chotpitayasunondh T, Sangsajja C, Chuchottaworn C, Farrar J, Puthavathana P. Influenza A viral loads in respiratory samples collected from patients infected with pandemic H1N1, seasonal H1N1 and H3N2 viruses. Virol J. 2010;7:75. doi: 10.1186/1743-422X-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fouchier RA, Bestebroer TM, Herfst S, Van Der Kemp L, Rimmelzwaan GF, Osterhaus AD. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J Clin Microbiol. 2000;38:4096–4101. doi: 10.1128/jcm.38.11.4096-4101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 39.Killian ML. Hemagglutination assay for the avian influenza virus. Methods Mol Biol. 2008;436:47–52. doi: 10.1007/978-1-59745-279-3_7. [DOI] [PubMed] [Google Scholar]
- 40.Ananthanarayan R, Paniker CK. Non-specific inhibitors of influenza viruses in normal sera. Bull World Health Organ. 1960;22:409–419. [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel U, Scholtissek C. Inhibition of the intracellular transport of influenza viral RNA by actinomycin D. Arch Virol. 1995;140:1715–1723. doi: 10.1007/BF01384336. [DOI] [PubMed] [Google Scholar]
- 42.Vreede FT, Ng AK, Shaw PC, Fodor E. Stabilization of influenza virus replication intermediates is dependent on the RNA-binding but not the homo-oligomerization activity of the viral nucleoprotein. J Virol. 2011;85:12073–12078. doi: 10.1128/JVI.00695-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dovas CI, Papanastassopoulou M, Georgiadis MP, Chatzinasiou E, Maliogka VI, Georgiades GK. Detection and quantification of infectious avian influenza A (H5N1) virus in environmental water by using real-time reverse transcription-PCR. Appl Environ Microbiol. 2010;76:2165–2174. doi: 10.1128/AEM.01929-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vester D, Lagoda A, Hoffmann D, Seitz C, Heldt S, Bettenbrock K, Genzel Y, Reichl U. Real-time RT-qPCR assay for the analysis of human influenza A virus transcription and replication dynamics. J Virol Methods. 2010;168:63–71. doi: 10.1016/j.jviromet.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 45.Nayak DP, Hui EK, Barman S. Assembly and budding of influenza virus. Virus Res. 2004;106:147–165. doi: 10.1016/j.virusres.2004.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ha SD, Ham B, Mogridge J, Saftig P, Lin S, Kim SO. Cathepsin B-mediated autophagy flux facilitates the anthrax toxin receptor 2-mediated delivery of anthrax lethal factor into the cytoplasm. J Biol Chem. 2010;285:2120–2129. doi: 10.1074/jbc.M109.065813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura T, Sunryd JC, Hebert DN. Sorting things out through endoplasmic reticulum quality control. Mol Membr Biol. 2010;27:412–427. doi: 10.3109/09687688.2010.495354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zinchuk V, Grossenbacher-Zinchuk O. Quantitative colocalization analysis of fluorescence microscopy images. Curr Protoc Cell Biol. 2014;62(Unit 4 19):11–14. doi: 10.1002/0471143030.cb0419s62. [DOI] [PubMed] [Google Scholar]
- 49.Liu C, Ma Y, Yang Y, Zheng Y, Shang J, Zhou Y, Jiang S, Du L, Li J, Li F. Cell Entry of Porcine Epidemic Diarrhea Coronavirus Is Activated by Lysosomal Proteases. J Biol Chem. 2016;291:24779–24786. doi: 10.1074/jbc.M116.740746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Von Magnus P. Propagation of the PR8 strain of influenza A virus in chick embryos. III. Properties of the incomplete virus produced in serial passages of undiluted virus. Acta Pathol Microbiol Scand. 1951;29:157–181. doi: 10.1111/j.1699-0463.1951.tb00114.x. [DOI] [PubMed] [Google Scholar]
- 51.Marriott AC, Dimmock NJ. Defective interfering viruses and their potential as antiviral agents. Rev Med Virol. 2010;20:51–62. doi: 10.1002/rmv.641. [DOI] [PubMed] [Google Scholar]
- 52.Yu WC, Chan RW, Wang J, Travanty EA, Nicholls JM, Peiris JS, Mason RJ, Chan MC. Viral replication and innate host responses in primary human alveolar epithelial cells and alveolar macrophages infected with influenza H5N1 and H1N1 viruses. J Virol. 2011;85:6844–6855. doi: 10.1128/JVI.02200-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pattnaik AK, Brown DJ, Nayak DP. Formation of influenza virus particles lacking hemagglutinin on the viral envelope. J Virol. 1986;60:994–1001. doi: 10.1128/jvi.60.3.994-1001.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meier-Ewert H, Compans RW. Time course of synthesis and assembly of influenza virus proteins. J Virol. 1974;14:1083–1091. doi: 10.1128/jvi.14.5.1083-1091.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshimura A, Kuroda K, Kawasaki K, Yamashina S, Maeda T, Ohnishi S. Infectious cell entry mechanism of influenza virus. J Virol. 1982;43:284–293. doi: 10.1128/jvi.43.1.284-293.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takizawa N, Momose F, Morikawa Y, Nomoto A. Influenza A Virus Hemagglutinin is Required for the Assembly of Viral Components Including Bundled vRNPs at the Lipid Raft. Viruses. 2016:8. doi: 10.3390/v8090249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 58.Hussain M, Galvin HD, Haw TY, Nutsford AN, Husain M. Drug resistance in influenza A virus: the epidemiology and management. Infect Drug Resist. 2017;10:121–134. doi: 10.2147/IDR.S105473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burster T, Giffon T, Dahl ME, Bjorck P, Bogyo M, Weber E, Mahmood K, Lewis DB, Mellins ED. Influenza A virus elevates active cathepsin B in primary murine DC. Int Immunol. 2007;19:645–655. doi: 10.1093/intimm/dxm030. [DOI] [PubMed] [Google Scholar]