Figure 2. Sequence comparison of human Pur proteins, Pura, Purb and two forms of Purg.
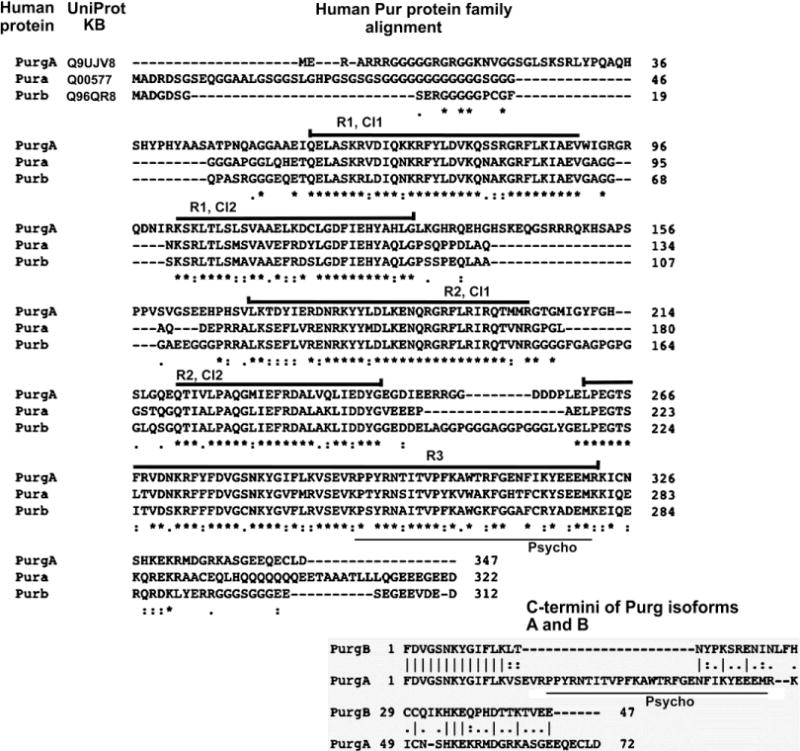
All four of the human protein Pur family members are aligned using the Clustal Omega algorithm. Pura, Purb and Purg-A are each derived from a distinct gene locus. Their entire sequences are aligned at top. Stars indicate amino acid identities; one dot indicates a substitution; two dots indicate a conservative substitution. Purg-B is transcribed from the Purg-A gene locus. Transcription of Purg-B, however, passes through the Purg-A stop point producing a 38 kb intron that is spliced out to make Purg-B mRNA. It is only the C-termini that differ between Purg-A and Purg-B. Sequences of these are presented at the bottom. Vertical lines indicate identities, and 2 dots indicate conservative substitutions. The Psycho motif is lightly underlined in both top and bottom alignments. In Pura it binds the Rb protein, among others (Ma et al., 1994; Johnson et al., 1995), and it is conserved in evolution. It is eliminated in Purg-B. Class 1 (Cl1) and Class2 (Cl2) segments of Pura repeats (R) 1, 2 and 3 are as indicated and heavily underlined.
