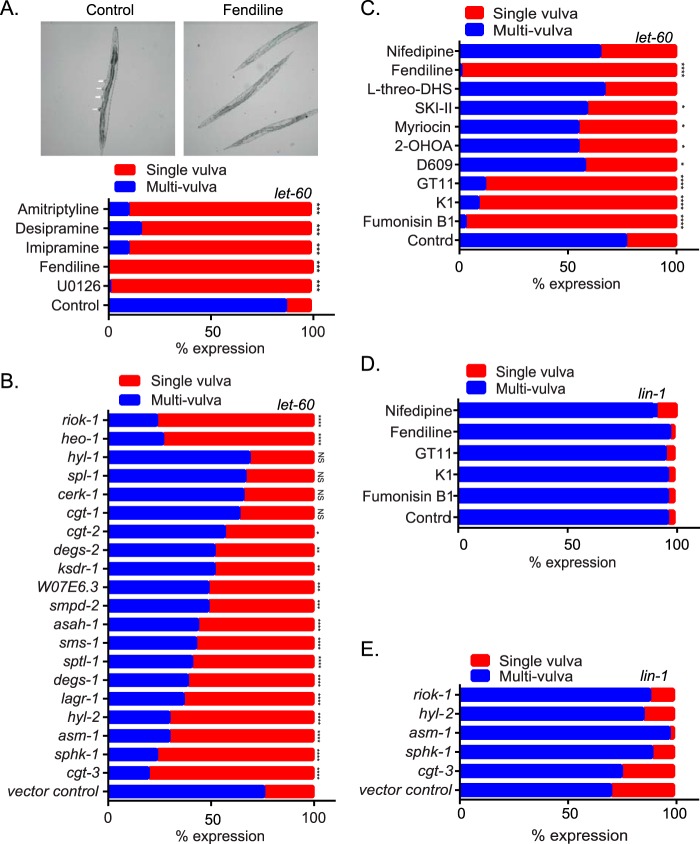
FIG 5.
Genetic and pharmacological perturbation of Cer-SM biosynthetic pathway alters K-Ras function in C. elegans. (A) Strain let-60(n1046) L1 larvae were cultured in M9 buffer containing the E. coli strain OP50 in the presence of vehicle (DMSO) or 30 μM fendiline, imipramine, desipramine, amitriptyline, or U0126. After 4 to 5 days, when worms reached the adult stage, they were scored for the presence of the multivulva phenotype using differential interference contrast (DIC)/Nomarski microscopy. Representative images of worms with the multivulva phenotype (shown by arrowheads) and suppression of this phenotype with fendiline are shown. (B) RNAi was induced by feeding let-60(n1046) L1 worms through adult stage with E. coli strain HT115, producing dsRNA to target genes. The presence of the multivulva phenotype was scored using DIC/Nomarski microscopy. (C) Strain let-60(n1046) L1 larvae were cultured in M9 buffer containing E. coli strain OP50 in the presence of vehicle (DMSO), 20 μM fumonisin B1, 50 nM myriocin, 100 μM D609, 175 μM 2-OHOA, 30 μM l-threo-DHS, 30 μM K1, 10 μM GT11, or 10 μM nifedipine. After 4 to 5 days, when worms reached the adult stage, they were scored for the presence of the multivulva phenotype using DIC/Nomarski microscopy. (D) lin-1(sy254) strain L1 larvae were cultured in M9 buffer containing the E. coli strain OP50 in the presence of vehicle (DMSO) or 30 μM fendiline, 20 μM fumonisin B1, 30 μM K1, 10 μM GT11, or 10 μM nifedipine. After 4 to 5 days, when worms reached the adult stage, they were scored for the presence of the multivulva phenotype using DIC/Nomarski microscopy. (E) RNAi was induced by feeding lin-1(sy254) L1 worms through adult stage with E. coli strain HT115, producing dsRNA to target genes. The presence of the multivulva phenotype was scored using DIC/Nomarski microscopy. One hundred to 200 worms were assayed per treatment or RNAi knockdown. Significant differences were evaluated in Student's t tests (*, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant).