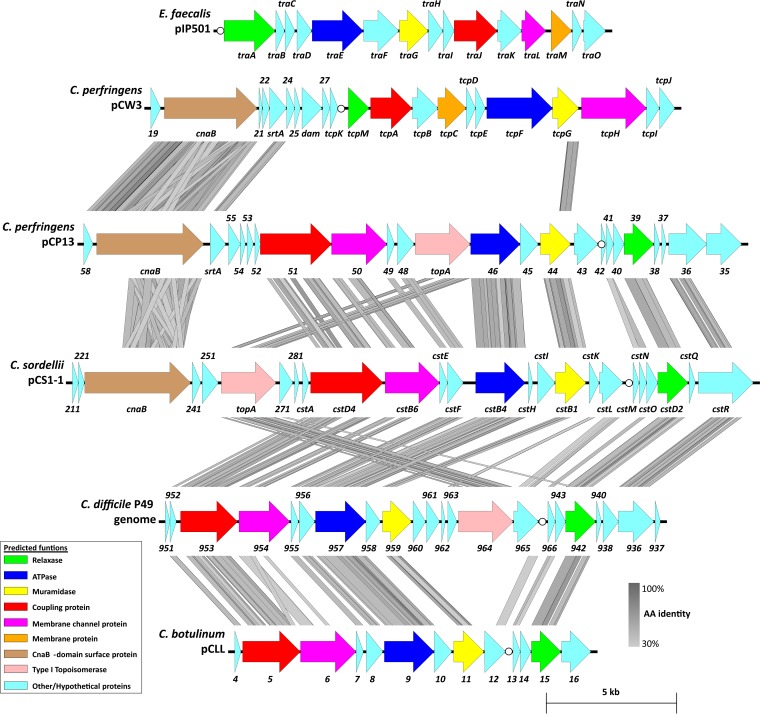
FIG 3 .
Comparison of the pCS1-1 cst locus and surrounding regions with both characterized and putative conjugation loci. Shown is a tblastX alignment of the characterized conjugation locus of broad-host-range plasmid pIP501 (AJ505823, L39769) and C. perfringens plasmid pCW3 (DQ366035), along with the putative conjugation locus from C. perfringens pCP13 (AP003515), C. sordellii pCS1-1 (LN679999), C. botulinum pCLL (CP001057), and the C. difficile strain P49 draft genome sequence (AVMN01000016). ORFs are colored according to known/predicted function (refer to key). Predicted oriT sequences are shown as a small white circle. Regions of amino acid identity between sequences are represented by gray bars. The cutoff value for amino acid identity is 30% across a region of 50 residues, with a maximum E value of 0.001. The higher the level of identity, the darker the gray, as illustrated by the legend. Previously named genes have been labeled as such. ORFs without classical gene names have been assigned a number designation. ORFs within the putative C. sordellii cst conjugation locus have been assigned a letter on the basis of the order in which they appear; however, those encoding putative components of a conjugation apparatus have been assigned a suffix based on the corresponding functional homologue from the Vir system of A. tumefaciens Ti plasmids (e.g., the VirD4 functional homologue is CstD4). E. faecalis, Enterococcus faecalis. The figure was produced using EasyFig.