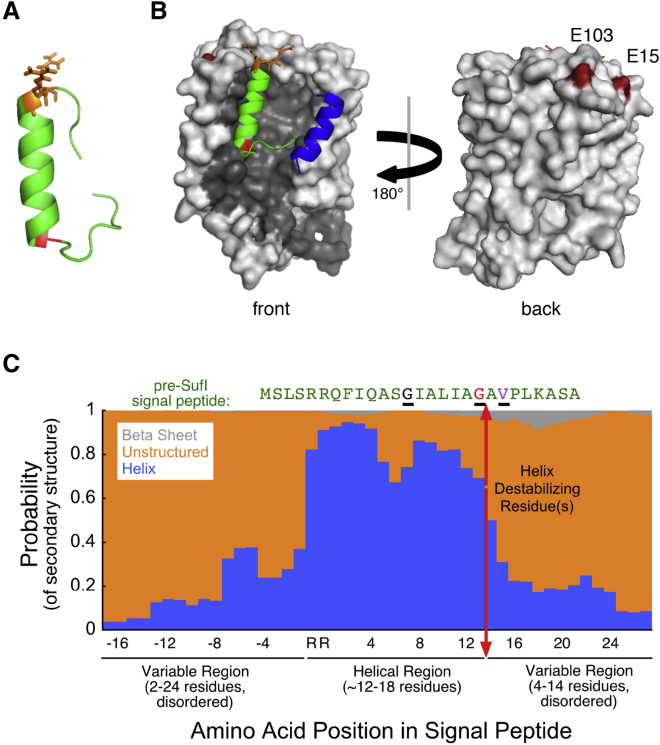
Figure 6.
Secondary structure of Tat signal peptides. (A) Shown here is the predicted secondary structure of the pre-SufI signal peptide (residues 1–27). The PEP-FOLD peptide structure prediction algorithm (48) yielded an h-region helix connected to an unstructured C-terminal region by a glycine (G) helix-breaking residue (red). The RR-motif (orange) is found at the N-terminal end of the helix. (B) Shown here is the pre-SufI signal peptide docked into the TatBC complex. Using ZDOCK (52), the TatB membrane domain (blue; residues L7-G21) was first positioned onto the TatC structure (gray; SWISS-MODEL: P69423) based on previous data indicating interactions with TM5 of TatC (12, 26). The pre-SufI signal peptide structure from (A) was then docked into this TatBC complex (lowest energy interaction) without any conformational relaxation of the Tat proteins or the signal peptide. The resultant signal peptide/receptor complex structure is consistent with the hairpin insertion by the TatBC complex. The RR-motif is near the two glutamic acid residues (dark red; E15 and E103) at the top of TatC involved in binding this motif, the tip of the hairpin is near the center of the bilayer, and the C-terminal end of the signal peptide is near TatB (see later cross-linking studies and the Discussion). (C) Shown here is the predicted secondary structures for 512 Tat signal sequences. The JPred structural propensity algorithm (50) predicts that the RR-motif and h-domain are largely helical, and that this central region of Tat signal peptides is flanked by unstructured domains. One or more helix-destabilizing residues (see text) at the C-terminal end of the h-domain helix is consistent with the bend necessary to form a hairpin. Note that some signal peptides have one or more helix-destabilizing residues within the h-domain (e.g., G13 of pre-SufI; black) before the helix-destabilizing residue(s) at the end of the helical domain (e.g., G19 of pre-SufI; red) (see also Fig. S6). V21, which deeply penetrates the membrane (Fig. 3) when the pre-SufI signal peptide is bound to TatBC, is identified in purple. Black underlines help to identify these three residues in the pre-SufI sequence.