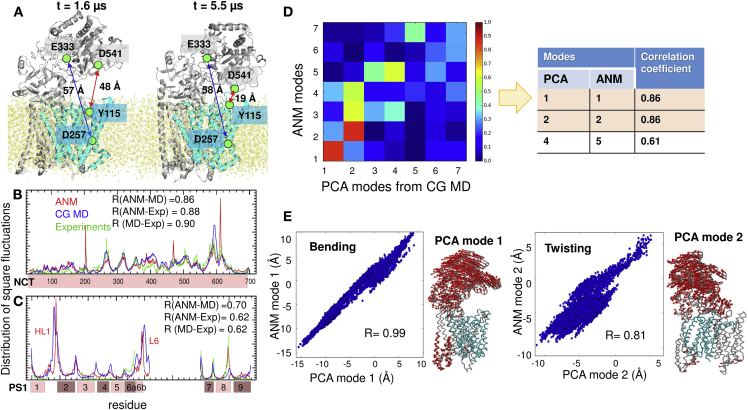
Figure 3.
Results from CG MD simulations of γ-secretase. (A) Motions of NCT with respect to PS1 (cyan). Two snapshots at t = 1.6 and 5.5 μs are displayed (from run 1). The distances between Asp257 and Glu333 (blue arrows) and Tyr115 and Asp541 (red arrows) are indicated. See Fig. S3 for more details. (B) Comparison of the MSF profile of NCT residues predicted by the ANM (red curve), observed in CG MD (blue), and deduced from ensemble analysis of five PDB structures (green). Correlations between the three pairs vary in the range 0.86–0.90, as indicated. (C) Same as (B), for PS1. (D) Correlations between the softest (seven) modes predicted by the ANM and those obtained from the PCA of 10 μs MD trajectory (run 1). High correlations are shown in dark red and weak correlations in dark blue. The table lists the pairs that exhibit the highest correlations. (E) Projections of 10,000 frames from 10 μs trajectory onto the ANM mode 1 and PCA mode 1 directions (left), and ANM mode 2 and PCA mode 2 directions (right). PCA mode 1 is equivalent to the bending (mode 1) predicted by ANM, as illustrated in the ribbon diagram, and PCA mode 2 is equivalent to the twisting mode (ANM mode 2). Similar results obtained in CG MD run 2 are presented in Fig. S2.