Figure 8.
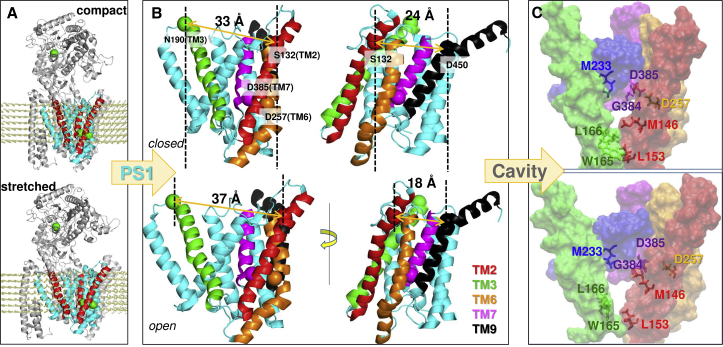
Asymmetric breathing motion of PS1. (A) A pair of conformers, compact and stretched, sampled during reconfiguration of PS1 along ANM mode 14 of the protease complex-lipid system is shown. The conformers are obtained based on an RMSD of 4 Å each with respect to the initial (PDB) structure. TM2, TM3, and TM7 are colored red. Glu333 in NCT1, and Asp257 and Asp385 in PS1, are shown in green spheres. (B) Close-up view of interhelical distance changes in the two conformers. TM2, TM3, TM6, TM7, and TM9 are colored as in Fig. 1C; other TM helices are cyan. The left diagrams display PS1 from the same perspective as in (A); the right diagrams are rotated around the normal to the membrane plane, to facilitate the visualization of the TM2–TM9 distance. Asp257, Asp385, Ser132 (TM2), Asn190 (TM3), and Asp450 (TM9) are shown in spheres colored by the corresponding TMs. S132–N190 (left) and S132–D450 (right) distances show opposite changes, i.e., an increase in the former is accompanied by a decrease in the latter and vice versa. (C) Catalytic cavity in the respective conformers, with TM2–TM7 shown in space-filling. Side chain positions obtained by energy minimization are shown for selected residues. See also Movie S3.