Figure 2.
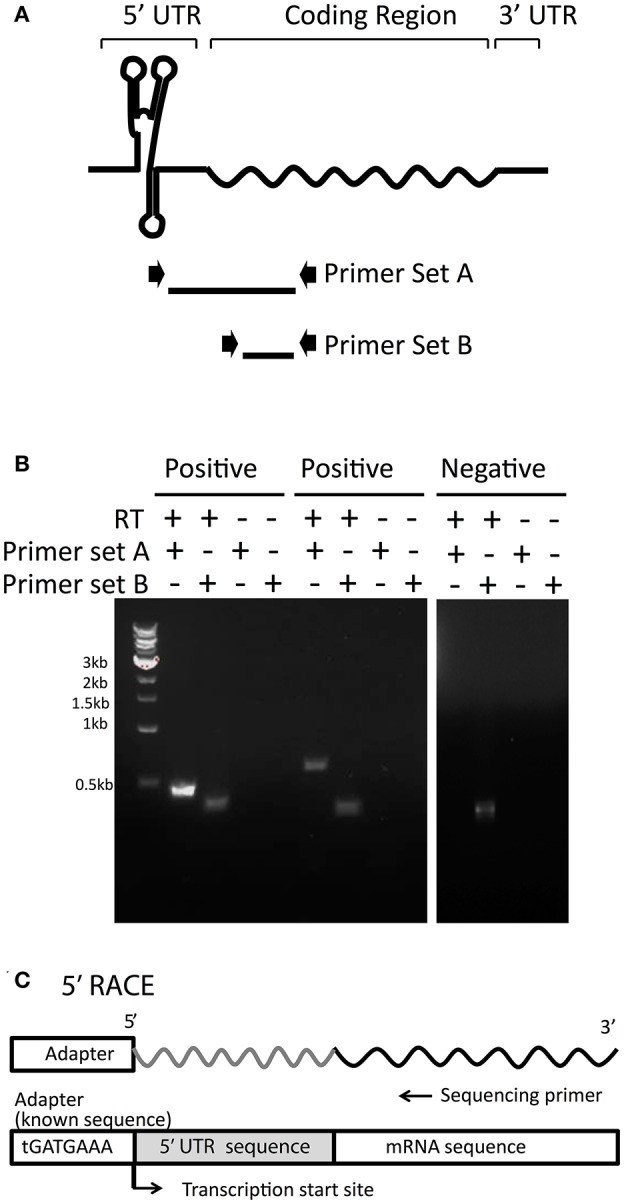
RT-PCR analysis to experimentally confirm 5′UTR candidates. (A) To confirm the 5′UTRs detected in transcriptomic data are actually transcribed with their downstream mRNAs, two sets of primers were designed. Primer Set A amplifies from the middle of the predicted UTR into the adjacent mRNA coding region. Primer Set B, as a control, amplifies the coding region only, indicating the amplification level of the transcript. (B) Examples of positive (confirmed) and negative (undetected) 5′UTR candidates are shown. RNA was reverse-transcribed and then subjected to PCR with Primer Set A and Primer Set B for each 5′UTR candidate. As a negative control, RNA was left without reverse transcription to indicate background levels of any residual DNA that could be left undigested from DNaseI treatment. Samples were then visualized on agarose gels, as shown in the examples. (C) For each 5′UTR confirmed by RT-PCR, 5′RACE was performed to determine the transcription start site by attaching an adapter of known sequence to the 5′ end and sequencing from the mRNA coding region upstream toward the adapter.
