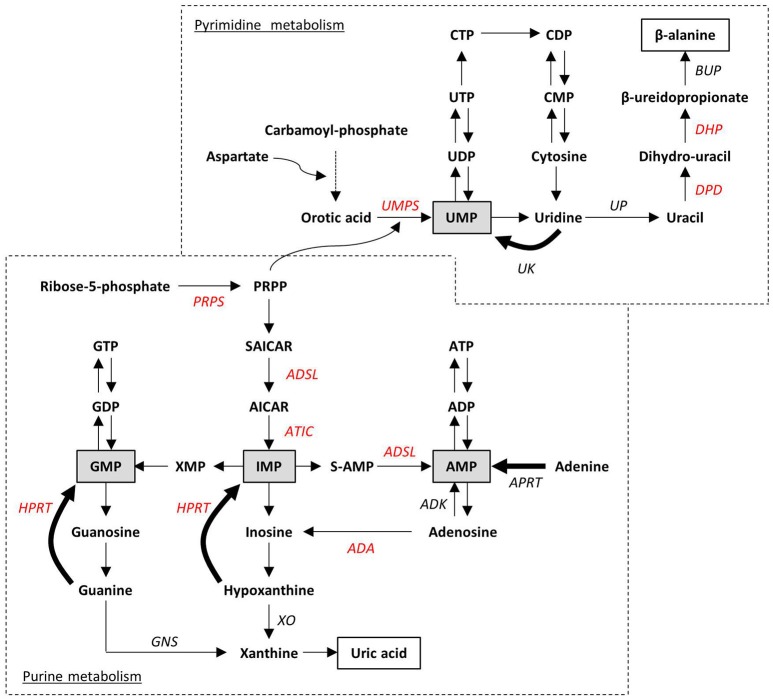
Figure 3.
Schematic representation of purine and pyrimidine metabolism. Ribose-5-phosphate and carbamoyl-phosphate are the starting points of the two de novo biosynthesis pathways. Salvage pathways are indicated with bold arrows. Monophosphate intermediates, representing the link between de novo biosynthesis and salvage pathways, are highlighted in gray boxes. End-products of purine and pyrimidine catabolism (i.e., uric acid and β-alanine) are in white boxes. Impairment at any enzymatic step can lead to inborn disorders. Enzymes whose altered functions have already been associated to the neurodevelopmental disorders described in the text are indicated in red. ADA, Adenosine deaminase; ADK, Adenosine kinase; ADSL, adenylosuccinate lyase; APRT, adenine phosphorybosyl-transferase; ATIC, AICA-ribotidetransformylase/IMP cyclohydrolase; BUP, β-ureidopropionase; DHP, dihydropyrimidinase; DPD, dihydropyrimidine dehydrogenase; GNS, guanase; HPRT, hypoxanthine-guanine phosphorybosyl-transferase; PRPP, 5-phosphorybosyl-1-pyrophosphate; PRPS, PRPP synthetase; S-AMP, adenylosuccinate; SAICAR: succinylaminoimidazole carboxamide ribotide; XO, xanthine oxydase; UK, uridine kinase; UMPS, UMP synthetase; UP, uridine phosphorylase.