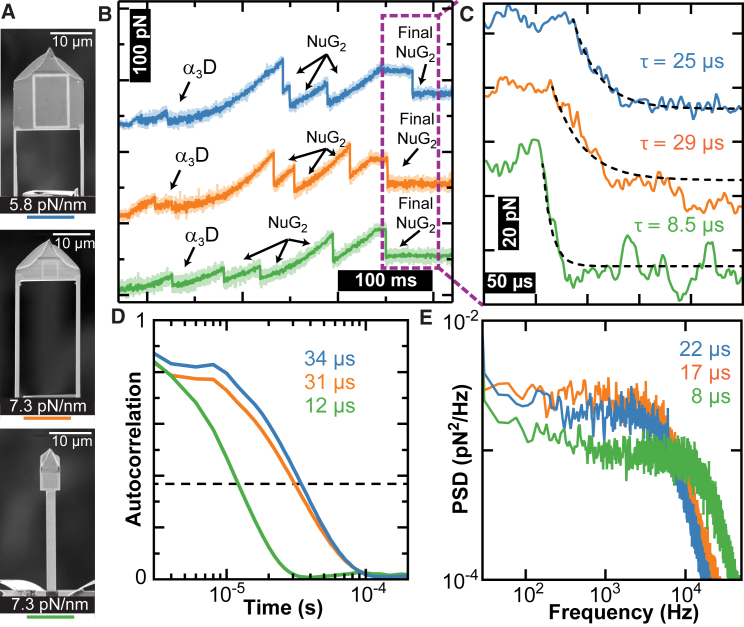
Figure 2.
Temporal resolution of different cantilever geometries. (A) Scanning electron microscopy images of the modified cantilevers. (B) Force-versus-time traces show unfolding of the (NuG2)2-α3D-(NuG2)2 construct. In this assay, the construct was stretched until the α3D and three NuG2 domains unfolded. The stage was further retracted until the polyprotein was held at ∼80 pN. The stage retraction was then stopped and the last folded NuG2 domain unfolded. Data smoothed to 2 kHz. (C) High-bandwidth force-versus-time traces from (B) showing the unfolding of the fourth NuG2 domain at v = 0 nm/s. Time constants determined from exponential fits. Data acquired at 500 kHz. (D) Autocorrelation of the cantilever motion after the final NuG2 domain unfolded but was still attached to the polyprotein. Time constants shown were determined from the 1/e point of the autocorrelation (dashed line). (E) Force PSDs of the 500-kHz data after unfolding of the final NuG2 domain. Time constants estimated from τ ≈ Q/(πƒc) based on the characteristic frequency, ƒc.