Figure 6.
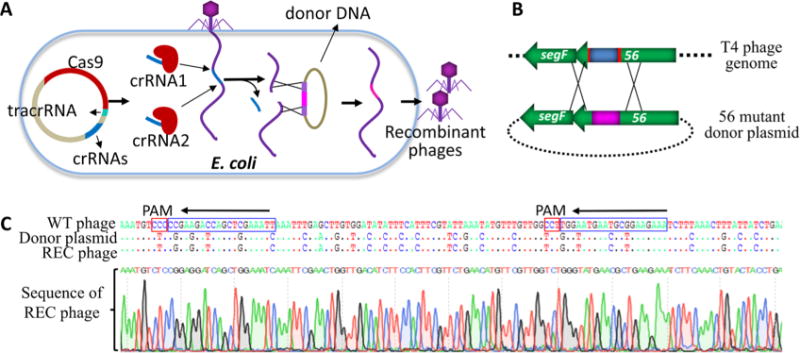
Genome editing using two adjacent spacers. (A) Schematic of gene replacement by CRISPR-Cas editing using two spacers. (B) Protospacers in phage T4 genome are shown in red and the region targeted for replacement is shown in blue. The two CRISPR-Cas9 complexes formed with the two different crRNAs will create breaks in the T4 genome at the corresponding protospacer sequences in g56. This results in excision of the intervening sequence. Recombination of the genome ends with the donor DNA flanking the inset will lead to replacement of the original sequence with that present in the donor DNA (shown in pink). (C) Sequencing of a random plaque showing the transfer of the new sequence into phage genome as a result of CRISPR-Cas mediated genome editing. Protospacer and PAM sequences are marked with blue and red boxes, respectively.
